Abstract
Background
This study was designed as a quasi-experiment to evaluate automatic inoculation of fecal specimens, using the automated specimen inoculator Previ Isola (bioMérieux, France).
Methods
We evaluated the quality of cultures, recovery rates of enteropathogenic bacteria (Salmonella, Shigella, Campylobacter, and Yersinia species), and cost-effectiveness in terms of technical time. The Previ Isola recovery rates for the two-year period from August 2009 to July 2011 were compared with historical manual inoculation data of the previous two years (August 2007 to July 2009). The regional (Baden-Württemberg) and nationwide (Germany) trends of recovery rates for this four-year period were referred.
Results
A total of 5,884 fecal specimens were collected over the study period. Most positive cultures were for Salmonella, followed by Campylobacter. Compared with the historical data, the numbers of Campylobacter-positive specimens for a year between August and July were increased significantly, from 19 in 2007-2008 and 10 in 2008-2009 to 32 in 2009-2010 (P=0.002) and 32 in 2010-2011 (P=0.003), respectively. During the study period, the official data for our region and nationwide did not show this increase in the recovery rate of Campylobacter. For Salmonella, Shigella, and Yersinia, no significant changes were observed. Compared with manual inoculation, the mean hands-on time with Previ Isola inoculation was significantly shortened, from 37:30 min to 8:42 min per 15 fecal specimens.
The rapid identification of microorganisms and subsequent susceptibility testing are highly important for a routine clinical microbiological laboratory [1]. On the one hand, most clinical laboratories are annually experiencing a rising number of specimens and requested analyses on a daily basis. On the other hand, the rapid diagnosis of an infection is essential for effective patient management [2].
Automation in clinical laboratories is on-going, with the aim to accelerate the detection of infectious agents. Various automation systems for microbiological purposes have recently been introduced into the market [3, 4]. Until recently, only a few automated specimen streakers were evaluated clinically [4, 5, 6]. The advantages of automated systems over manual inoculation range from a good reproducibility of cultures and a greater number of isolated colonies [1, 7] to potential cost savings when integrated into the laboratory work flow [6].
Fecal specimens are frequently sent to a clinical laboratory for the diagnosis of gastroenteritis by enteropathogenic bacteria or antibiotic-associated diarrhea due to Clostridium difficile. The gold standard for diagnosing gastroenteritis by enteropathogenic bacteria is still stool culturing for Salmonella, Shigella, Yersinia, or Campylobacter species.
The microbiological recovery of enteropathogenic bacteria requires time-consuming plate inoculation and mostly further subcultures to be able to isolate and identify suspected strains. The quality of plate inoculation is therefore important in a specimen like fecal specimens, which are full of aerobic and anaerobic bacteria. Economically, cost-effectiveness can be monitored by savings both in agar plate consumption and in the technical time as a surrogate marker.
In our institution in Heidelberg University Hospital, the number of fecal specimens has steadily increased over the years, and a system for automated specimen inoculation (Previ Isola system; bioMérieux, Marcy l'Etoile, France) was therefore validated and introduced into routine diagnosis. Furthermore, in our institution, we plate screening swabs and urine cultures with the help of automation.
In this study, we prospectively evaluated automated specimen inoculation for fecal specimens over a two-year study period. The manual-loop method, which was performed beforehand, was exchanged with plate inoculation with the help of Previ Isola. The change of methodology was carried out throughout the entire study period. We evaluated the quality of cultures, recovery rates of enteropathogenic bacteria, and cost-effectiveness in terms of technical time. Results of culture were compared against historical data as well as official data from health authorities for the whole of Germany and our specific region (Baden-Württemberg).
The Previ Isola system (bioMérieux) was developed for the automated and standardized inoculation and streaking of plates. Using a circular applicator, a standard quantity of inoculum is deposited every time and is pressure-control streaked on agar plates. One hundred eighty agar plates can be stored in five input cassettes to deliver a sufficient loading capacity for the quick processing of specimens. The streaking of at least 180 plates per hour guarantees a high standard of plate processing. The Previ Isola system can be used not only for liquid specimens, but also for swab systems with transport media, such as liquid Amies medium, to improve the diagnosis of aerobes, ananaerobes, fastidious bacteria, and fungi [8].
The study was conducted at the 2,000-bed tertiary care Heidelberg University Hospital from August 2009 until July 2011. During the two-year study period, fecal specimens sent for microbiological analysis were collected in standardized containers and sent by pneumatic tube within a few minutes for examination. The duration of transport did not change during the study. The patient demographics and mix remained equal during the study duration. The total number of positive cultures was defined as the non-duplicated number of patients (only one specimen per episode of gastroenteritis) within the observational period of six months.
Specimens were plated onto Columbia agar with 5% sheep blood (Becton Dickinson, Franklin Lakes, NJ, USA), xylose-lysine-desoxycholate (XLD) agar, Yersinia selective agar (CIN), and Campylobacter agar (bioMérieux). Before plating by the Previ Isola system, 1 g of stool was suspended in 1.5 mL of 0.45% saline in a 5 mL polystyrene tube (Sarstedt, Nürnbrecht, Germany). An 18 µL aliquot of this suspension was plated. For Salmonella and Shigella species, in parallel to the saline inoculation, a selenite enrichment broth was inoculated manually with a loopful of fecal specimen.
The comparison of methodologies involved comparing the Previ Isola automated specimen inoculation of fecal specimens with those specimens inoculated via the manual-loop method. This was to validate that via automated specimen inoculation, there is no loss in the detection limit of enteric pathogens. One hundred manually plated fecal specimens spiked with serial dilutions of enteric pathogens (gold standard) were compared with plating by the Previ Isola system during routine work time. Colonies were counted after 24 hr and 48 hr.
One hundred consecutive fecal specimens were plated in parallel by the manual-loop method and Previ Isola inoculation. The technical staff judged the quality of the automated streaking using the following criteria: the morphology of colonies, the number of isolated colonies per plate, and their personal impression of the automatic method as being superior, equal, or inferior to the manual method.
Plates with Columbia agar and XLD agar were incubated at 36 1℃ in 5% CO2 for 18-24 hr. The CIN agar plates were incubated at 28±1℃ for 48 hr. The Campylobacter agar plates were incubated at 42±1℃ in the presence of a microaerophilic generator for 48 hr. The inocula in enrichment broth were incubated at 36±1℃ for 18-24 hr. The growth conditions were in accordance with standard German protocols [9]. In the absence of growth of enteric pathogen on the solid media after 24 hr, one drop of the broth was plated onto XLD agar and incubated overnight at 36±1℃.
Plates were reviewed at 24 hr and 48 hr. Colonies were identified according to standard microbiological procedures, using the Vitek 2 system (bioMérieux) and a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Bruker Daltonics, Billerica, MA, USA). The plates were examined by different and independent members of the technical and medical staff.
Results from the study period were compared with two collections of data, described below.
i) Historical set of data: 5,140 fecal specimens were collected in the two years before the study period and had been manually plated in a three-quadrant method by experienced technical personnel (manual loop-to-plate method [10 µL]).
ii) Official data from the German health authority: microbiological laboratories in Germany are obliged to notify cultures of Salmonella species, Shigella species, Campylobacter species, and Yersinia species immediately. Data are collected at the Robert Koch Institut, Berlin, Germany, and can be queried at http://www3.rki.de/SurvStat. Furthermore, data are transmitted to the European Centre for Disease Prevention and Control (ECDC), Stockholm, Sweden.
The mean time taken to manually plate 1 and 15 fecal specimens in a three-quadrant method was calculated for three different members of the technical staff during the study period. Likewise, the mean time for the automatic inoculation of 1 and 15 fecal specimens using the Previ Isola system was also calculated.
Chi square analysis was used for statistical analyses. P values <0.05 were regarded as significant.
Only growth on solid media was taken into account. Growth on solid media after cultivation on enrichment media was omitted for statistical purposes. Data from the two-year study period were compared with the historical data from the previous two years. All data were analyzed using SPSS 19.0 (SPSS Inc., Chicago, IL, USA).
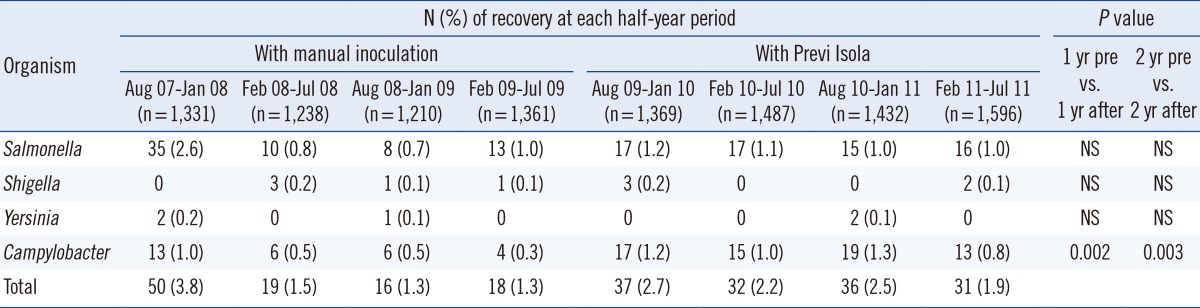
A total of 5,884 fecal specimens were collected from patients over the study period. The total numbers of positive cultures for Salmonella, Shigella, Yersinia, and Campylobacter species are shown half-yearly in Table 1.
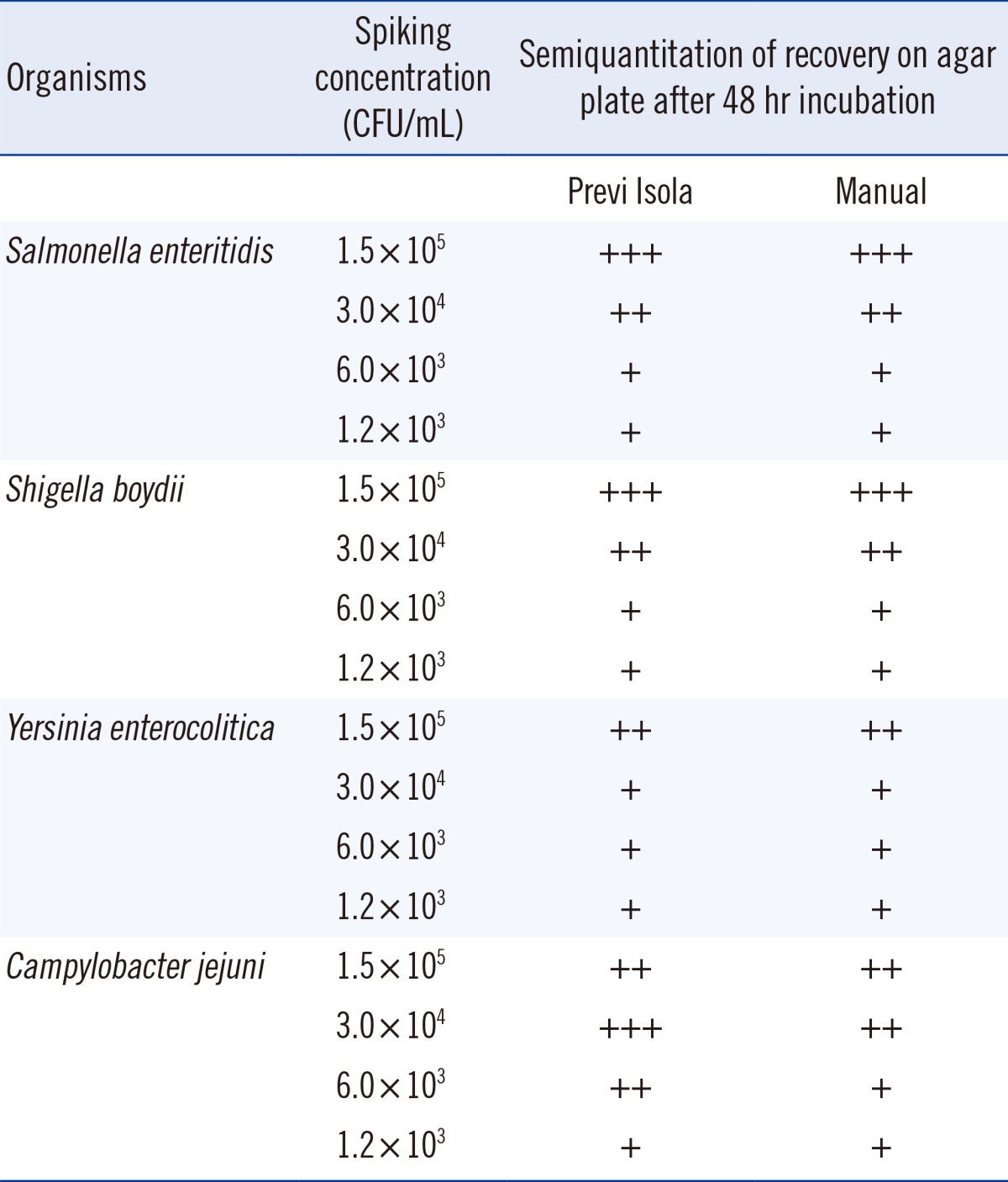
Serial dilutions of enteric pathogens in fecal specimens showed complete agreement between the two methods of inoculation (Table 2). The inter-rate agreement between the Previ Isola automated specimen inoculation and manual inoculation was determined with the help of kappa (κ) scores, as described previously (data not shown) [6].
Colony growth on agar plates inoculated by Previ Isola or manually is shown in Fig. 1. The suitability of the automated streaking was judged by the technical staff as being "superior" to manual plating in 48%, and as "equal" in another 48%. Only in 4% was "inferior" documented by the technicians. Automatically streaked agar plates allowed for better processing of colonies in most cases, as colonies were more often single-standing and less confluent. Different microorganisms could be distinguished more easily.
Fig. 1 shows that the morphology of colonies and the number of single-standing and less-confluent colonies were better after automated plate inoculation than by manual-loop plating.
Results of the two-year study period were compared with those taken in the two years before the start of the study. The numbers of positive cultures in the prior two years inoculated with the manual-loop method are shown in Table 1.
The absolute numbers of our study were compared with the official data for the whole of Germany and for our region, Baden-Württemberg (Fig. 2). The data shown comprise the study period and the historical period of two years before.
The mean time to plate one fecal specimen was 0:35 min for Previ Isola and 2:35 min for the technical staff. The mean time to plate 15 fecal specimens was 8:42 min for Previ Isola and 37:30 min for the technical staff. The time difference with regard to 15 specimens was 28:48 min.
Chi square analysis revealed a significant increase of Campylobacter detection rates in the one-year period after the introduction of the Previ Isola system, compared with the one-year period before its introduction (P<0.002). The same could be shown when comparing the two-year period before the introduction of the Previ Isola system with the two-year period afterwards (P=0.003). For Salmonella, Shigella, and Yersinia species, no significant statistical differences could be found when comparing these same periods before and after Previ Isola introduction. These data are shown in Table 1.
Automation in microbiological laboratories leads to more efficient and accurate processing [3, 5, 10]. Nevertheless, studies that compare automated systems with manual inoculation are still lacking for many types of specimens. In our institution, wound swabs have been extensively evaluated, with a favorable outcome [6]; co-workers could show that the Previ Isola system is able to reduce the time and cost of broncho-pulmonary specimen management [11], but other specimen types such as fecal specimens have not been investigated so far.
To the best of our knowledge, this is the first study to evaluate the performance of an automated inoculation system for fecal specimens in a routine clinical microbiological laboratory. This study was intended to evaluate the performance of the Previ Isola system for fecal specimens with regard to quality of cultures, recovery rates of enteropathogenic bacteria, and the potential of cost savings.
Single-standing colonies may lead to savings in terms of reduced agar plate consumption, and consequently to savings in terms of technical time. Our data showed that the savings in technical time with the help of automated plate inoculation can be enormous. Interestingly, however, even if the cost-effectiveness of modern microbiological diagnostics is shown [12], it is still unclear whether a quicker and better microbiological diagnosis is reflected in the clinical outcome of patients and has an influence on parameters like length of stay or mortality.
The streaker showed high reliability and stability. No cross-contamination between different specimens was detected. Prior to the study, we excluded that automated specimen inoculation leads to a loss of detection of pathogens.
The numbers of reported cases for the whole of Germany indicate an overall decrease in the number of Salmonella cases and an increase in the number of Campylobacter cases. Campylobacteriosis is the most commonly reported gastrointestinal disease in the European Union [13], whereas in the US, FoodNet reported that the incidence of Campylobacter infections from 1996 to 2005 had declined so much that Salmonella infections currently outnumber those caused by Campylobacter. The incidence of Campylobacter infections in Germany is about 80/100,000 and is significantly higher than that of salmonellosis [14]. However, the national trend is not reflected in the regional data (Baden-Württemberg) where our institution is located. In our region, a significant increase of Campylobacter cases from 2007 until 2011 was not observed. After consultation with our infection control team and communications with our regional health authority (Rhine-Neckar district), we excluded any possibility of enteric pathogen clustering due to an outbreak during the study period. When comparing the number of our positive cultures for Campylobacter with historical data, one can see a clear increase, reflecting the rising numbers of campylobacteriosis monitored by the ECDC [15]. However, when comparing our numbers of Campylobacter detection rates with the regional and national figures from the health authorities, it almost appears as though there are disproportionately higher numbers of positive cultures for Campylobacter since adopting the change of methodology. Nevertheless, we cannot get to clear conclusions when comparing the two methods and different specimens and in different times. The higher recovery rate of Campylobacter from Previ Isola inoculation was significant. As shown in the spiked specimens of 103-104 colony-forming unit (CFU)/mL, isolation using the Previ Isola system resulted in a higher quantity of certain inocula than the manual-loop method, as well as more single-standing colonies with a larger size. This effect of Previ Isola inoculation was observed with only Campylobacter in the experiment with the spiked specimens. Therefore, automatic inoculation using Previ Isola may guarantee a better recovery of fastidious species like Campylobacter, which is often covered by the overgrowth of normal flora of fecal specimens.
The incidence of salmonellosis in Germany is about 30/100,000 [14]. In our institution, Salmonella species are still the most frequently detected pathogens in stool cultures in contrast to ECDC data [15]. With the introduction of the Previ Isola system, there was no increase in the detection rate for Salmonella species. We therefore exclude fluctuations in the instances of pathogen recovery. Other works have even shown a decrease in the number of positive cultures for Salmonella species owing to on-farm control measures, better refrigeration, consumer education, and better food handling in restaurants and homes [16]. Whether the decline in Salmonella detection rates relative to historical data is due to epidemiological circumstances or to the improved inoculation by the Previ Isola system remains to be determined. Automated plate inoculation has several advantages over classical microbiological diagnostics. One such advantage is that it can be used easily and delivers a standardized inoculation. The time to plate 15 fecal specimens was significantly different when comparing manual and automatic inoculations. Our results are consistent with former findings that analyzed time saving for the plating of urine specimens [17].
Automation also enables precise inoculation and better and faster processing, and no specific technical experience is necessary. The main limitation to our study was the lack of a control group during the study period. When comparing the two inoculation methodologies, we succeeded in showing that automated specimen inoculation does not affect the detection of microorganisms negatively.
For reasons of feasibility and costs, a parallel design was not possible, as that would have meant testing more than 5,000 specimens in parallel over two years of study in a routine clinical university microbiological laboratory. Therefore, the historical and official data from health authorities served as the control groups. Since we are one of the largest university microbiological laboratories in Germany, with heterogeneous inpatient and outpatient clinics, our submissions differ from private commercial laboratories that work on specimens of mainly non-hospitalized patients.
Our study demonstrates that inoculation by Previ Isola guarantees an improvement in the routine culture of fecal specimens, with better sensitivity for Campylobacter and less hands-on time. Furthermore, the quality of culture, recovery rates of organisms, or potential cost savings in plate consumption are likely influenced. Further prospective parallel studies of manual and automated inoculation methods are needed to prove our hypothesis.
Notes
References
1. Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J Clin Microbiol. 2008; 46:1281–1284. PMID: 18272700.

2. Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Rev Infect Dis. 1983; 5:54–70. PMID: 6828812.

3. Bourbeau PP, Ledeboer NA. Automation in clinical microbiology. J Clin Microbiol. 2013; 51:1658–1665. PMID: 23515547.

4. Bourbeau PP, Swartz BL. First evaluation of the WASP, a new automated microbiology plating instrument. J Clin Microbiol. 2009; 47:1101–1106. PMID: 19158259.

5. Mutters NT, Hodiamont CJ, de Jong MD, Overmeijer HP, van den Boogaard M, Visser CE. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann Lab Med. 2014; 34:111–117. PMID: 24624346.

6. Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J Clin Microbiol. 2012; 50:2732–2736. PMID: 22692745.

7. King GW, Kath GS, Siciliano S, Simpson N, Masurekar P, Sigmund J, et al. Automated agar plate streaker: a linear plater on Society for Biomolecular Sciences standard plates. J Biomol Screen. 2006; 11:704–711. PMID: 16844965.

8. Van Horn KG, Audette CD, Tucker KA, Sebeck D. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn Microbiol Infect Dis. 2008; 62:471–473. PMID: 18814991.

9. Kist M, Bockemühl J, Aleksic S, Altwegg M, Autenrieth IB, Bär W, et al. Infektionen des Darms. In : Mauch H, Lütticken R, editors. Qualitätsstandards in der mikrobiologisch-infektiologischen Diagnostik. München, Jena: Urban & Fischer;2000. p. 32–39.
10. Tilton RC, Ryan RW. Evaluation of an automated agar plate streaker. J Clin Microbiol. 1978; 7:298–304. PMID: 348722.

11. Nebbad-Lechani B, Emirian A, Maillebuau F, Mahjoub N, Fihman V, Legrand P, et al. New procedure to reduce the time and cost of broncho-pulmonary specimen management using the Previ Isola® automated inoculation system. J Microbiol Methods. 2013; 95:384–388. PMID: 24184016.

12. Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012; 50:3301–3308. PMID: 22855510.

13. Fitzenberger J, Uphoff H, Gawrich S, Hauri AM. Urban-rural differences of age- and species-specific campylobacteriosis incidence, Hesse, Germany, July 2005-June 2006. Euro Surveill. 2010; 15:pii: 19693.

14. Robert-Koch-Institut. Epidemiologie der Kryptokokkose in Deutschland von 2004 bis 2010. Epid Bull. 2012; 29:275. http://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2012/Ausgaben/29_12.pdf?__blob=publicationFile.
15. European Centre for Disease Prevention and Control (ECDC). Annual epidemiological report. Reporting on 2011 surveillance data and 2012 epidemic intelligence data. Stockholm: ECDC;2013.
16. Marcus R, Rabatsky-Ehr T, Mohle-Boetani JC, Farley M, Medus C, Shiferaw B, et al. Dramatic decrease in the incidence of Salmonella serotype Enteritidis infections in 5 FoodNet sites: 1996-1999. Clin Infect Dis. 2004; 38(S3):S135–S141. PMID: 15095182.
17. Funke G, Kunert T, Barth B, Fulchiron C, Bossy G. New automated solution for plate streaking: comparative evaluation of the PREVI Isola in a microbiology lab. 19th European Congress of Clinical Microbiology and Infectious Diseases. Abstract number, P887. Helsinki: 2009. Updated on May 2009. http://www.lbtinnovations.com/assets/Uploads/eccmid092l.pdf.
Fig. 1
Colony growth on Campylobacter agar streaked manually (left) and on corresponding agar plates streaked by the Previ Isola system (right).
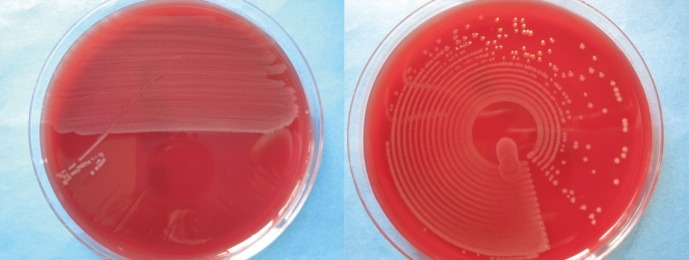
Fig. 2
Trends of reported cases of Salmonella, Shigella, Yersinia, and Campylobacter over four years, from the official data for Germany and for the Baden-Württemberg region. Number of cases for the pathogens indicated in the given time period are shown for Germany (A) and Baden-Württemberg (B). Source: Robert Koch Institut; http://www3.rki.de/SurvStat.
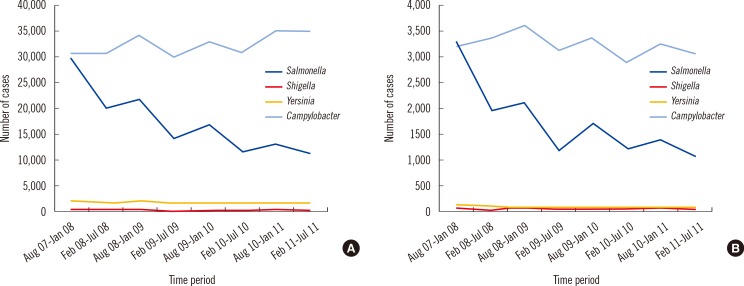
Table 1
Comparison of recovery rates of enteropathogens from stool cultures for a one-and two-year-period before and after introduction of automatic inoculation using Previ Isola




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download