Abstract
Background
By conventional methods, the identification of anaerobic bacteria is more time consuming and requires more expertise than the identification of aerobic bacteria. Although the matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems are relatively less studied, they have been reported to be a promising method for the identification of anaerobes. We evaluated the performance of the VITEK MS in vitro diagnostic (IVD; 1.1 database; bioMérieux, France) in the identification of anaerobes.
Methods
We used 274 anaerobic bacteria isolated from various clinical specimens. The results for the identification of the bacteria by VITEK MS were compared to those obtained by phenotypic methods and 16S rRNA gene sequencing.
Results
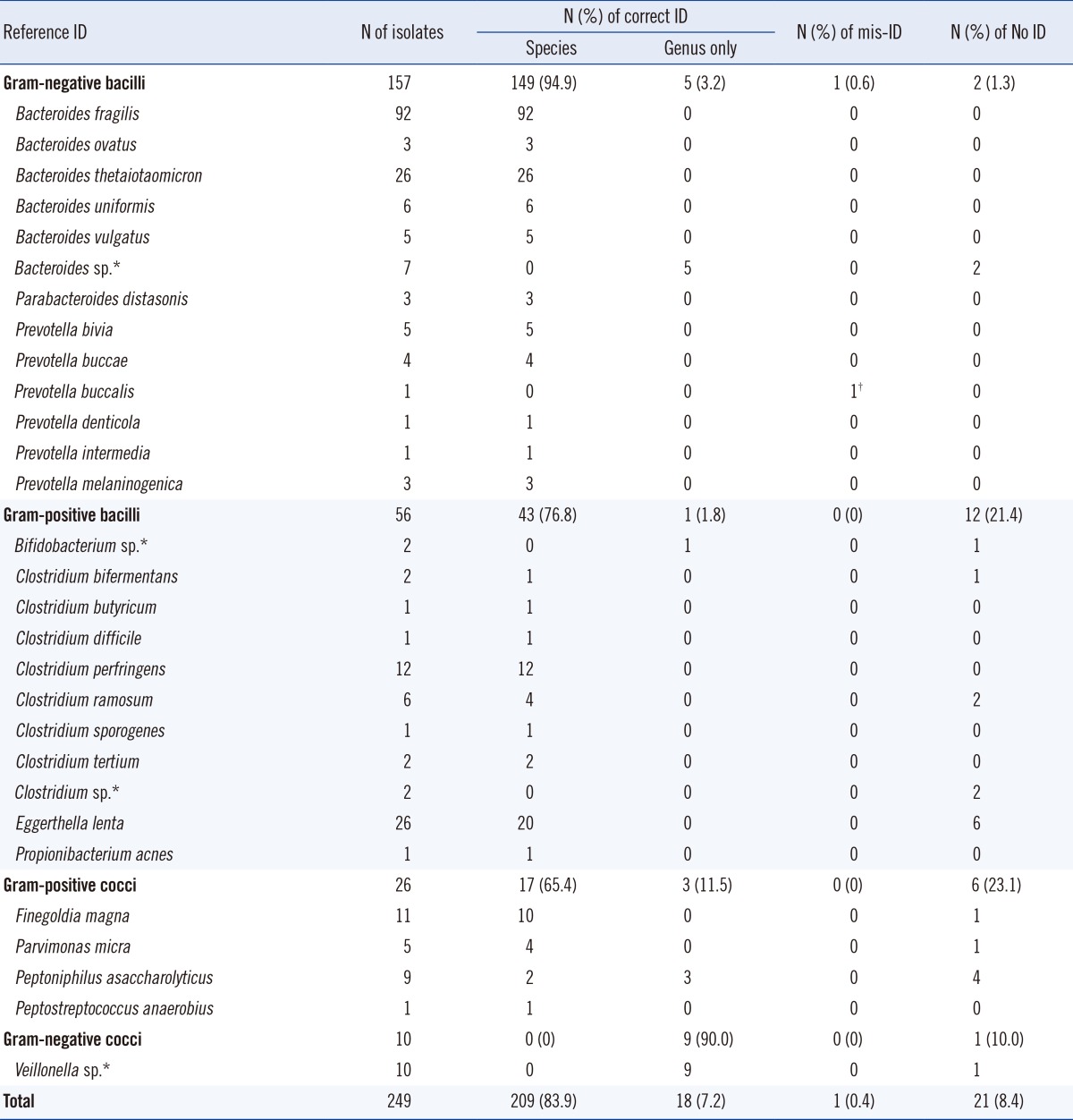
Among the 249 isolates included in the IVD database, the VITEK MS correctly identified 209 (83.9%) isolates to the species level and an additional 18 (7.2%) at the genus level. In particular, the VITEK MS correctly identified clinically relevant and frequently isolated anaerobic bacteria to the species level. The remaining 22 isolates (8.8%) were either not identified or misidentified. The VITEK MS could not identify the 25 isolates absent from the IVD database to the species level.
Go to : 
It is well known that many anaerobic bacteria play a significant role in clinical infections. However, many clinical microbiology laboratories cannot satisfactorily identify anaerobic bacteria [1, 2]. Phenotypic methods for the identification of anaerobic bacteria, including the currently popular commercial identification systems, are laborious and often unreliable for the identification of rare species [2].
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) is a technique used to analyze the mass-to-charge ratio of various molecules that are ionized from crystallized sample materials by laser pulses [3]. MALDI-TOF MS has recently been used for the identification of microorganisms in clinical laboratories. The microbes are identified by comparing the protein spectra from the MALDI-TOF MS to those in a reference database. Although some published papers reporting the evaluation of commercially available MALDI-TOF MS systems showed inconsistent results because of differences in the evaluation protocols, it is apparent that the technology is more accurate than conventional phenotypic methods for routine bacterial identification [4, 5, 6, 7, 8, 9, 10]. Most errors were attributed to an incomplete spectral database or the inability of the MS spectra to differentiate species that were similar. Among the microorganisms, species of anaerobes were frequently not identified, probably because of the lack of reference spectra in the databases [11].
We evaluated the VITEK MS (bioMérieux, Marcy-l'Etoile, France) for the identification of isolated anaerobic bacteria from clinical specimens in comparison to the identification of the bacteria by Gram staining, morphology testing, using commercial biochemical systems, and 16S rRNA gene sequencing.
Go to : 
Clinical isolates of 274 anaerobic bacteria were recovered from clinical specimens including normally sterile body fluids including blood (n=83), peritoneal fluids (n=84), pleural fluids (n=11), and tissue samples (n=12; e.g., skin, bone) from patients at a university hospital in South Korea in 2011. The bacteria were identified at isolation by conventional phenotypic methods including Gram staining, cell morphology tests, and the API Rapid ID 32A and/or VITEK 2 ANC card (bioMérieux) systems. The isolates were kept frozen at -80℃ until species identification by the VITEK MS was performed.
All frozen isolates were subcultured twice on Brucella blood agar plates (Asan Pharm, Hwaseong, Korea) for 48 hr at 35℃ in an anaerobic chamber (Forma, Marietta, OH, USA) for identification by the MALDI-TOF MS system. A single colony or multiple small colonies of each isolate were smeared onto the VITEK MS-DS target slide (bioMérieux), supplied in a 48-well microscope slide format, and divided into 3 acquisition groups of 16 spots each using a 1-µL disposable loop (SPL Life Sciences, Pocheon, Korea). The prepared samples were covered with 1 µL of α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (bioMérieux) and dried at room temperature. The mass spectra were acquired using a VITEK MS Axima Assurance mass spectrometer (bioMérieux). The isolates were identified using the Advanced Spectrum Classifier (ASC) algorithm, which compares the obtained spectra with the typical spectra of each organism in the VITEK MS in vitro diagnostic (IVD) 1.1 database (which includes more than 25,000 spectra covering 585 species). For system calibration and internal identification control, Escherichia coli ATCC 8739 was used. The result from the first test with the VITEK MS, which provided a single choice at species level with ≥60% confidence, was used. If the test provided two or more choices, a single choice at the species level with <60% confidence, or no identification, it was immediately repeated with other colonies from the same agar plates. The final identification was then made after comparison with the reference identification.
The results of bacterial identification obtained by methods such as Gram staining, morphology tests, and commercial biochemical systems were used as the reference. However, when the commercial systems failed to identify the species or in the case of discordant results when compared to the VITEK MS, partial 16S rRNA gene sequencing was performed for identification of the bacteria. We performed 16S rRNA gene sequencing of a PCR product sized approximately 800 bp using the universal primers 4F (5'-TTG GAG AGT TTG ATC CTG GCT C-3') and 801R (5'-GGC GTG GAC TTC CAG GGT ATC T-3'), and the sequences were analyzed using GenBank (http://www.ncbi.nlm.nih.gov) and EzTaxon [12]. The species identified with a ≥99% match by 16S rRNA gene sequencing were accepted according to the CLSI guideline MM18-A [13].
The species identified using the VITEK MS were defined as follows: (i) correct identification of species: results of identification by VITEK MS were identical to those obtained by the reference identification techniques at the species level; (ii) only the correct genus was identified: results of the identification by VITEK MS were identical at the genus level to those obtained by the reference identification techniques; (iii) no identification: VITEK MS failed to identify the bacteria; (iv) misidentification: results obtained by VITEK MS were different from those obtained by other reference identification techniques.
Go to : 
Among the 274 anaerobic bacterial isolates tested, data for the genus or species of 249 isolates (90.9%) were available in the VITEK MS IVD 1.1 database (Table 1). Therefore, the performance of the system for the identification of the 249 isolates was evaluated. Of these 249 isolates, 209 (83.9%) were correctly identified at the species level, and an additional 18 (7.2%) were identified only at the genus level. The system provided no identification and misidentification for 21 (8.4%) and 1 (0.4%) isolates, respectively.
Among the 157 gram-negative bacilli, 149 (94.9%) were correctly identified at the species level. All the isolates of Bacteroides fragilis (n=92) and Bacteroides thetaiotaomicron (n=26) tested, which were the most frequently isolated species, were correctly identified. Five isolates of Bacteroides sp. were correctly identified at the genus level, but two were not identified. One isolate of Prevotella buccalis was misidentified as Pseudoflavonifractor capillosus.
Among the gram-positive bacilli, 76.8% (43/56) of the isolates were correctly identified at the species level. All 12 isolates of Clostridium perfringens were correctly identified. However, among the 26 Eggerthellalenta isolates, the most frequently isolated gram-positive bacilli, 6 were not identified. In addition, 2 of the 6 Clostridium ramosum isolates, one Clostridium bifermentans, both the Clostridium spp., and one Bifidobacterium spp. were not identified.
Most of the gram-positive cocci such as Finegoldia magna (10/11), Parvimonas micra (4/5), and Peptostreptococcus anaerobius (1/1) were correctly identified to the species level. However, 4 of the 9 Peptoniphilus asaccharolyticus could not be identified. Among the 10 isolates of Veillonella spp., 9 were identified as Veillonella parvula, but one could not be identified.
In total, 38 isolates were identified at only the genus level, misidentified, or not identified by the commercial system. Therefore, 16S rRNA gene sequencing was required for reference identification of the following isolates: Bacteroide sfragilis (n=3), Bacteroides thetaiotaomicron (n=1), Bacteroides uniformis (n=3), Bacteroides spp. (n=7), Prevotella buccalis (n=1), Prevotella denticola (n=1), Prevotella melaninogenica (n=1), Bifidobacterium spp. (n=2), Clostridium ramosum (n=6), Clostridium spp. (n=2), Finegoldia magna (n=1), and Veillonella spp. (n=10). Among these isolates the VITEK MS correctly identified Bacteroides fragilis (n=3), Bacteroides thetaiotaomicron (n=1), Bacteroides uniformis (n=3), Prevotella denticola (n=1), Prevotella melaninogenica (n=1), Clostridium ramosum (n=4), and Finegoldia magna (n=1). Among the 7 Bacteroides spp. isolates, Bacteroides uniformis (n=1), Bacteroides ovatus (n=3), and Bacteroides thetaiotaomicron (n=1) were identified by the VITEK MS.
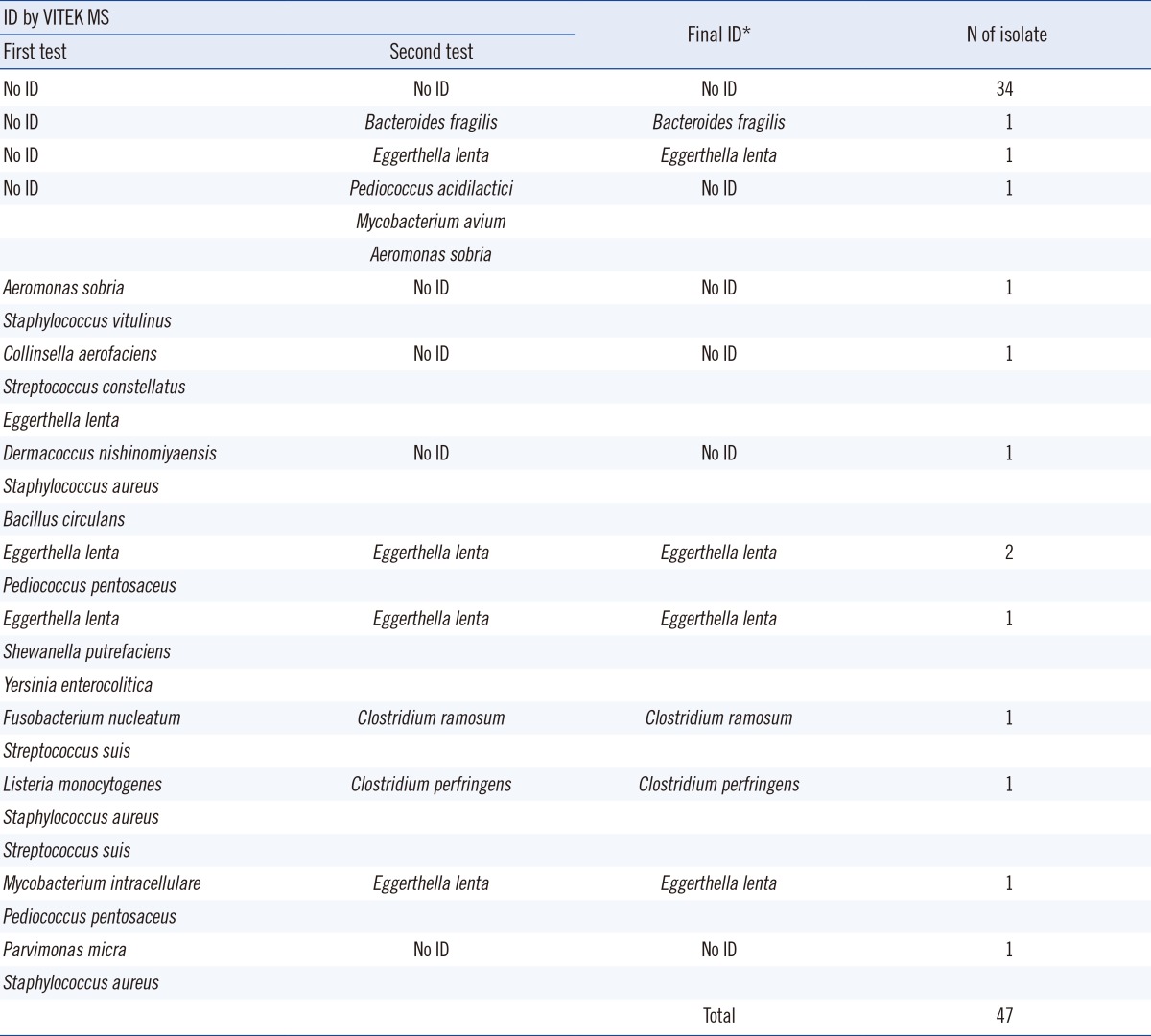
Among the 47 isolates that were retested by the VITEK MS, which included 10 isolates initially identified as 2 or more species and 37 isolates that were initially unidentified, species of 8 isolates were finally identified (Table 2).
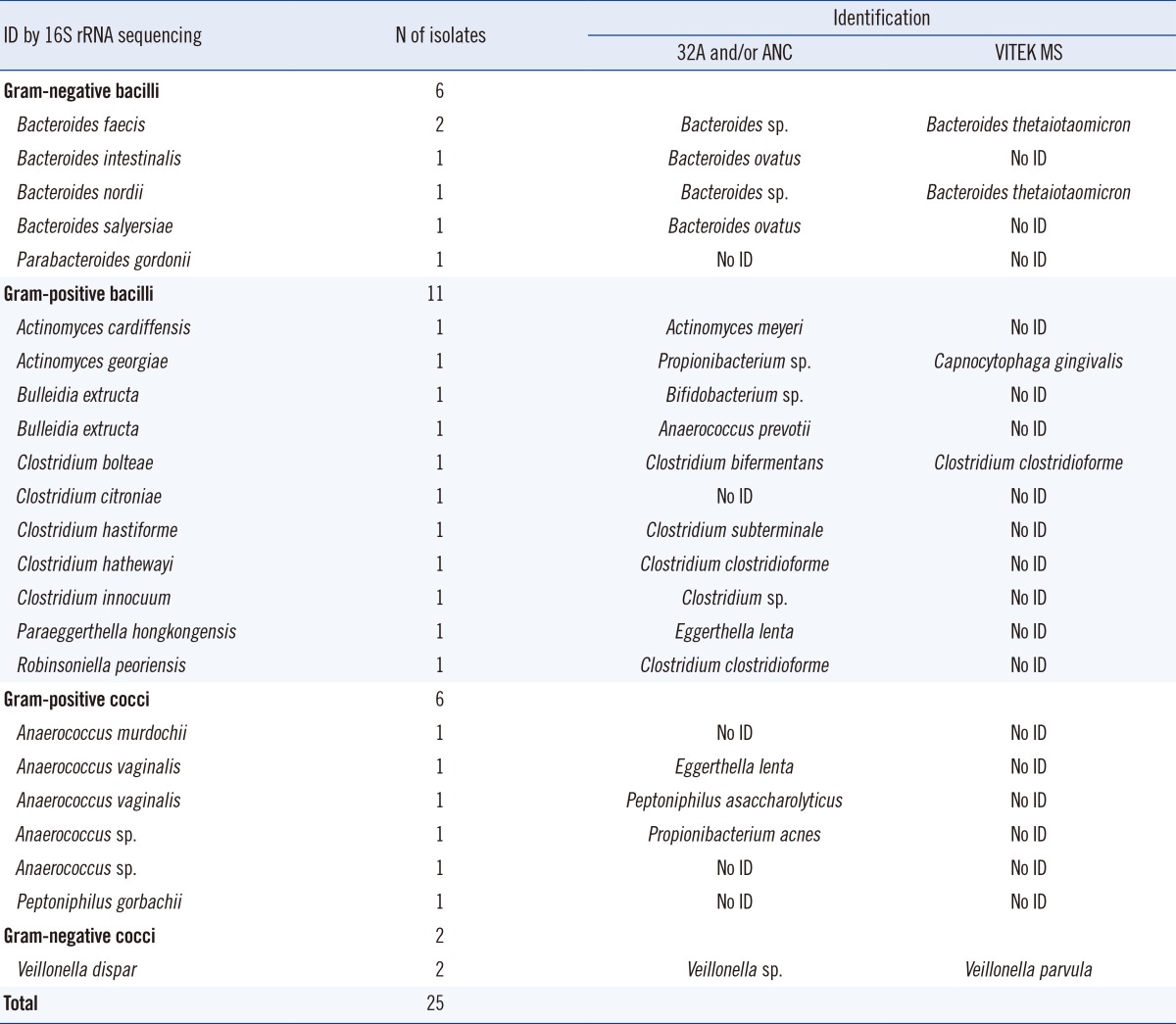
Among the 25 total isolates identified as a reference without a VITEK MS IVD 1.1 database (Table 3), only 6 isolates (24.0%; 2 Bacteroides faecis, 1 Bacteroides nordii, 1 Clostridium bolteae, and 2 Veillonella dispar) were correctly identified to the genus level by the VITEK MS, whereas other isolates were either not identified or misidentified. However, with the commercial biochemical systems, 12 isolates (48.0%; 6 Bacteroides spp., 4 Clostridium spp., 2 Veillonella dispar, and 1 Actinomyces cardiffensis) were correctly identified at the genus level.
Go to : 
Identification of microorganisms using MALDI-TOF MS gives several benefits over the phenotypic methods that are currently being used. Firstly, the species can be identified within a few minutes, and the reference spectra database can easily be modified and edited. In addition, the cost of running the VITEK MS is lower than that for other identification systems [3]. Moreover, MALDI-TOF MS is capable of identifying microorganisms from a single colony [3]. If sufficient growth is present in primary culture plates, microbiologists no longer have to perform subcultures or presumptive identifications using biochemical tests; therefore, MALDI-TOF MS is especially useful for the identification of anaerobic bacteria.
In this study, we used the VITEK MS with the IVD 1.1 database. Recently, the IVD 1.1 database of the VITEK MS was updated to the VITEK MS database 2.0 (29,873 spectra covering 755 species). In total, 170 bacterial and fungal organisms were added in the 2.0 version compared to the 1.1 version. However, because there was no change in the data of anaerobic bacteria, we considered that there would be no difference in the performance of the identification techniques for anaerobes made by using the two versions of the database. Among the 249 isolates with spectra present in the IVD 1.1 database, 209 isolates (83.9%) were identified at the species level. In particular, the VITEK MS provided accurate identification at the species level for the frequently isolated clinical bacteria, such as Bacteroides fragilis (92/92), Bacteroides thetaiotaomicron (26/26), Clostridium perfringens (12/12), and Finegoldia magna (10/11). However, 6 of the 26 Eggerthellal enta and 4 of the 9 Peptoniphilus asaccharolyticus were not identified by the VITEK MS because a sufficient number of mass peaks were not collected. These results were different from those of previous studies that were performed by using three MALDI-TOF MS systems (Bruker Biotyper, VITEK MS RUO, and VITEK MS IVD) [14, 15, 16]. Justesen et al. [14] showed that the Bruker Biotyper and VITEK MS RUO systems correctly identified 67.2% (195/290) and 49.0% (142/290) of isolates, respectively, at the species level. In another study, the VITEK MS IVD system showed a higher rate of species identification for 73 anaerobes than the Bruker system (75.3% and 61.6%, respectively) [15]. Recently, Garner et al. [16] evaluated the performance of the VITEK MS using VITEK MS database 2.0 for the identification of anaerobes, which showed a higher rate of correct species identification (91.2%) than ours. However, in contrast to our study, their study did not involve the evaluation of clinically relevant anaerobes. In particular, the genus Eggerthella, consisting of one of the frequently isolated anaerobes, for which no identification result was obtained for 6 of the 26 isolates used in our study, was not included in the study by Garner et al. [16]. Both the studies had the same limitation of the lack of several clinically important anaerobes such as the those belonging to the Fusobacterium and Propionibacterium genera. The differences in the rate of correct identification in each of these studies are believed to result from differences in the number of anaerobic bacteria included in each study.
MALDI-TOF MS showed better performance for the isolates present in the reference database than commercial biochemical systems, such as the VITEK 2 ANC card or the Rapid ID 32A. In total, 38 isolates could not be identified at the species level by the commercial system. Among these isolates, the VITEK MS correctly identified 14 isolates, which indicates that the VITEK MS performs better than the commercial phenotypic tests.
For accurate and reliable identification by MALDI-TOF MS, the quality and amount of reference spectra present in the database are important. The 25 isolates absent from the IVD 1.1 database could not be correctly identified. These isolates were rare species or species from newly described taxa. Therefore, the system's database should be further expanded and optimized for the species that were misidentified, and very rare species or recently named species should be added to the database for more accurate identification.
In conclusion, the VITEK MS showed good performance for the identification of most anaerobic bacteria isolated from clinical specimens. The VITEK MS is an easier and more reliable system for bacterial identification than commercial biochemical systems.
Go to : 
References
1. Murphy EC. Gram-positive anaerobic cocci-commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013; 37:520–553. PMID: 23030831.
2. Nagy E. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: a new possibility for the identification and typing of anaerobic bacteria. Future Microbiol. 2014; 9:217–233. PMID: 24571074.

3. Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol. 2012; 93:965–974. PMID: 22198716.

4. Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, et al. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J Clin Microbiol. 2012; 50:2034–2039. PMID: 22495566.

5. Culebras E, Rodríguez-Avial I, Betriu C, Gómez M, Picazo JJ. Rapid identification of clinical isolates of Bacteroides species by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Anaerobe. 2012; 18:163–165. PMID: 21963387.
6. Fedorko DP, Drake SK, Stock F, Murray PR. Identification of clinical isolates of anaerobic bacteria using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012; 31:2257–2262. PMID: 22371295.

7. La Scola B, Fournier PE, Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. 2011; 17:106–112. PMID: 21672636.

8. Nagy E, Becker S, Kostrzewa M, Barta N, Urbán E. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J Med Microbiol. 2012; 61:1393–1400. PMID: 22700545.

9. Jamal WY, Shahin M, Rotimi VO. Comparison of two matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry methods and API 20AN for identification of clinically relevant anaerobic bacteria. J Med Microbiol. 2013; 62:540–544. PMID: 23242640.

10. Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J Clin Microbiol. 2012; 50:2568–2576. PMID: 22593596.

11. Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013; 26:547–603. PMID: 23824373.

12. Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007; 57:2259–2261. PMID: 17911292.

13. Clinical and Laboratory Standards Institute. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. Approved guideline, MM18-A. Wayne, PA: Clinical and Laboratory Standards Institute;2008.
14. Justesen US, Holm A, Knudsen E, Andersen LB, Jensen TG, Kemp M, et al. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol. 2011; 49:4314–4318. PMID: 21998433.

15. Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012; 50:1313–1325. PMID: 22322345.

16. Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, et al. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK® MS system. Clin Microbiol Infect. 2014; 20:355–359. PMID: 23991748.

Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download