Dear Editor
MDS is a clonal disorder marked by ineffective hematopoiesis, cytopenias, clonal chromosomal abnormalities, and a variable predilection to undergo clonal evolution to AML. Multiple genetic aberrations occur during the clonal evolution of MDS, and in the majority of cases, somatic mutations result from the deletion of all or part of a chromosome [1].
BCR-ABL1 is a hybrid from the ABL1 gene on chromosome 9 and the BCR gene on chromosome 22. The fusion protein encoded from this gene has strong tyrosine kinase activity and is involved in the pathogenesis of several hematologic disorders. BCR-ABL1 genes are categorized into three types, based on differences in the BCR gene's breakpoint, which appear to be related to the disease phenotype. The BCR-ABL1 fusion gene is found in CML and in some cases of acute lymphoblastic leukemia [2]. However, reports on BCR-ABL1-positive MDS cases are extremely rare [3456]. Here, we report an unusual case of MDS progressing to AML with an e1a2 BCR-ABL1 fusion transcript and a complex karyotype, including monosomy 7.
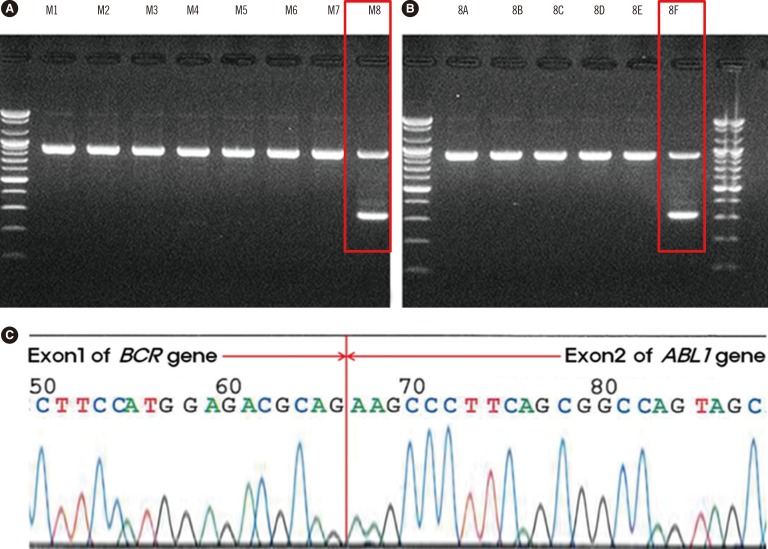
A 65-yr-old man was admitted for evaluation of persistent pancytopenia with 9% blasts in the peripheral blood. He presented with general weakness and breathlessness. Evaluation of the bone marrow (BM) disclosed hypercellularity with mild erythroid hyperplasia, blasts count of 18%, and remarkable dyshematopoiesis (Fig. 1). Immunophenotype analysis of BM cells using flow cytometry showed that the blasts were positive (>20% of cells) for CD13 (45%), CD33 (97%), CD117 (68%), CD34 (97%), and HLA-DR (43%), and negative for other megakaryocytic and lymphoid markers. Multiplex reverse transcriptase polymerase chain reaction (RT-PCR) evaluation of total RNA isolated from BM cells (HemaVision kit; DNA-Diagnostic, Risskov, Denmark) indicated an e1a2 (p190 BCR-ABL1) rearrangement, confirmed on two independent cDNAs and via direct sequencing (Fig. 2). Quantitative RT-PCR analysis (Real-Q BCR-ABL1 quantification kit; BioSewoom, Seoul, Korea) revealed the BCR-ABL1 to ABL1 transcript ratio to be 0.001339977. The fluorescence in situ hybridization signal for BCR-ABL1 rearrangement was found in one interphase cell out of 296 analyzed cells (Fig. 1D). There was no evidence of other molecular abnormalities such as mutations in JAK2 or calreticulin. Conventional cytogenetic analysis revealed a complex abnormality without the Philadelphia (Ph) chromosome: 44,XY,del(5)(q31),-7,del(12)(p12),-14,-16,+mar [cp20]. The final diagnosis was MDS subtype refractory anemia with excess blasts-2 (RAEB-2). The patient died before undergoing a follow-up BM examination owing to lung cancer, which was found shortly after the MDS diagnosis.
BCR-ABL1-positive de novo MDS is a very rare disease, and when it manifests with excess blasts, as in this case, it may be confused with the accelerated phase of CML. However, several features of this case differentiate it from CML. First, our patient did not have organomegalies, such as splenomegaly, or any related symptoms, which are common in CML. Second, our patient did not display basophilia in the peripheral blood or BM, which is common in CML. Third, BM examination showed severe dyshematopoietic features without findings of myeloproliferative neoplasms. Fourth, monosomy 7 and del(5)(q31) identified using chromosomal analysis are typical aberrations of MDS.
It is common that BCR-ABL1-negative MDS changes to BCR-ABL1-positive MDS after disease progression [3456]. In the case of BCR-ABL1-positive MDS, most patients show excess blasts on the verge of transformation to AML [678], which implies that the appearance of BCR-ABL1-positive clones is likely to be closely associated with leukemogenesis and triggers MDS to transform into AML.
Jacobsen et al. [9] proposed three possible explanations for the acquisition of the late-appearing Ph chromosome: 1) it may have been present on initial presentation; however, the techniques used were inadequate to detect the BCR rearrangement; 2) it may represent further evidence of multistep pathogenesis; and 3) the presence of the Ph chromosome in some cells may represent the development of a new clone of cells in leukemic relapse. In this case, we presume that the third explanation may be fitting. The BCR-ABL1-positive clone was most likely detected on emergence because the BCR-ABL1 copy number and BCR-ABL1 to ABL1 transcript ratio during quantitative RT-PCR analysis were found to be very low. Moreover, a typical Ph chromosome was not found in 20 metaphase cells by conventional cytogenetic analysis. To the best of our knowledge, this is the first report of de novo MDS RAEB-2 with the cryptic e1a2 subtype of BCR-ABL1 rearrangement and complex chromosomal abnormality in Korea. Currently, there is no established treatment method that is distinguished from the usual MDS treatment. In the future, a more systematic study of this disease is needed with respect to serial biologic monitoring, therapy, and outcome.
Acknowledgments
This study was supported by grants from the Leading Foreign Research Institute Recruitment Program of the NRF, the Korea Ministry of Education, Science and Technology (MEST) (No. 2011-0030034), and the National R&D Program for Cancer Control, Korea Ministry of Health & Welfare (No. 2013-1320070).
References
1. Lichtman MA. Myelodysplasia or myeloneoplasia: thoughts on the nosology of clonal myeloid diseases. Blood Cells Mol Dis. 2000; 26:572–581. PMID: 11112390.

2. Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996; 88:2375–2384. PMID: 8839828.
3. Kakihana K, Mizuchi D, Yamaguchi M, Sakashita C, Fukuda T, Yamamoto K, et al. Late appearance of Philadelphia chromosome with the p190 BCR/ABL chimeric transcript in acute myelogenous leukemia progressing from myelodysplastic syndrome. Rinsho Ketsueki. 2003; 44:242–248. PMID: 12784657.
4. Manabe M, Yoshii Y, Mukai S, Sakamoto E, Kanashima H, Nakao T, et al. Late appearing Philadelphia chromosome as another clone in a patient with myelodysplastic syndrome harboring der(5;12)(q10;q10) at diagnosis. Rinsho Ketsueki. 2012; 53:618–622. PMID: 22790637.
5. Fukunaga A, Sakoda H, Iwamoto Y, Inano S, Sueki Y, Yanagida S, et al. Abrupt evolution of Philadelphia chromosome-positive acute myeloid leukemia in myelodysplastic syndrome. Eur J Haematol. 2013; 90:245–249. PMID: 23240925.

6. Keung YK, Beaty M, Powell BL, Molnar I, Buss D, Pettenati M. Philadelphia chromosome positive myelodysplastic syndrome and acute myeloid leukemia-retrospective study and review of literature. Leuk Res. 2004; 28:579–586. PMID: 15120934.

7. Lesesve JF, Troussard X, Bastard C, Hurst JP, Nouet D, Callat MP, et al. p190bcr/abl rearrangement in myelodysplastic syndromes: two reports and review of the literature. Br J Haematol. 1996; 95:372–375. PMID: 8904895.
8. Tucker D, Hamilton MS, Kerr JP, Wickham C, Hunter H. Lytic bone disease as the presenting feature of Philadelphia-positive monosomy 7 myelodysplasia progressing to acute myeloid leukaemia. Gene. 2012; 501:219–221. PMID: 22537673.

9. Jacobsen RJ, Himoe E, Sacher RA, Shashaty GG. Late appearance of Philadelphia chromosome. Br J Haematol. 1986; 63:392–394. PMID: 3459549.

Fig. 1
Morphological and molecular cytogenetic analyses of bone marrow cells. A bone-marrow aspirate from the patient showed dyserythropoiesis (A), dysmegakaryopoiesis (B), and dysgranulopoiesis (C). Fluorescence in situ hybridization with a BCR-ABL1 dual-color dual-fusion probe demonstrated that most cells showed no genomic rearrangement. However, one interphase cell out of 296 analyzed cells harbored a BCR-ABL1 fusion signal (inner box). The ABL1 gene on chromosome 9 is labeled in red, and the BCR gene on chromosome 22 is labeled in green (D).
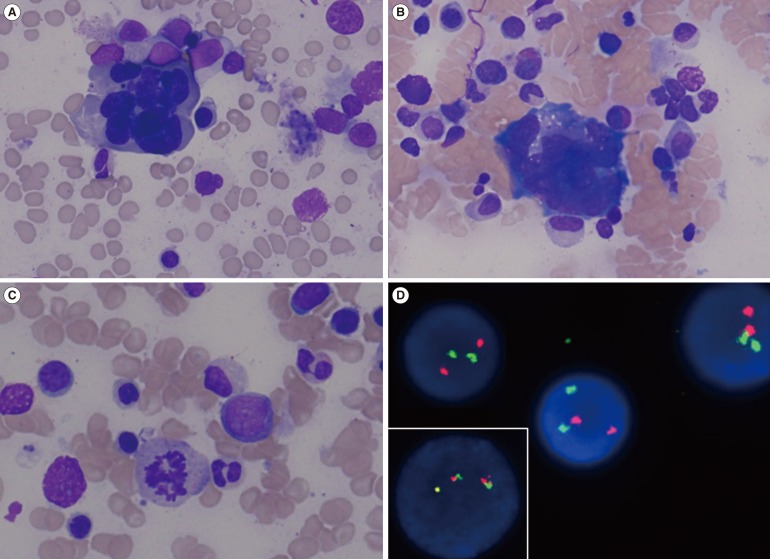
Fig. 2
Detection of p190 BCR-ABL1 fusion transcript by multiplex reverse transcriptase -PCR and direct sequencing. Analysis of RNA samples collected from the patient's bone marrow (A, B). The screening kit produced a single band in the M8 lane (A), and the split-out kit produced a single 320-base-pair band in the 8F lane (B), indicating the minor presence of the BCR-ABL1 fusion transcript (e1a2 type). Direct sequencing for the confirmation of a BCR-ABL1 gene rearrangement showed the fusion between exon 1 of the BCR gene and exon 2 of the ABL1 gene (C).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download