Abstract
Background
The present analysis focuses on phenotypic and genotypic characterizations of efflux-mediated erythromycin resistance in Streptococcus pneumoniae due to an increase in macrolide resistance in S. pneumoniae worldwide.
Methods
We investigated the prevalence of efflux-mediated erythromycin resistance and its relevant genetic elements from 186 specimens of S. pneumonia isolated from clinical and normal flora from Tehran, Iran. The presence of erythromycin resistance genes was tested by PCR with two sets of primers, specific for erm(B) and mef(A/E), and their genetic elements with tetM, xis, and int genes. Isolates were typed with the BOX PCR method and tested for resistance to six antibiotics.
Results
Antibiotic susceptibility tests revealed that 100% and 47% isolates were resistant to tetracycline and erythromycin, respectively. The erythromycin and clindamycin double-disc diffusion test for macrolide-lincosamide-streptograminB (MLSB) resistance phenotype showed 74 (84%) isolates with the constitutive MLSB phenotype and the remaining with the M phenotype. BOX PCR demonstrated the presence of 7 types in pneumococci with the M phenotype. Fourteen (16%) isolates with the M phenotype harbored mef(A/E), tetM, xis, and int genes.
Go to : 
Streptococcus pneumonia colonizes the human nasopharynx and is the leading causative agent of otitis media, respiratory infection, bacteremia, and meningitis, leading to high morbidity and mortality worldwide [1, 2]. In recent years, treatment for S. pneumoniae has become difficult owing to the global rise in the prevalence of antibiotic resistance, particularly against first-line antibiotics such as erythromycin and penicillin [3].
Macrolide resistance in S. pneumoniae is mediated by 2 major mechanisms: ribosomal modification by 23S rRNA methylation [4] and active drug efflux [5] encoded by erm(B) and mef genes, respectively. The macrolide efflux pump proteins belong to the major facilitator superfamily of transporters, and several mef genes have also been described in other microbial species [6].
M phenotype, which is mediated by mef genes, was first demonstrated in S. pneumonia and S. pyogenes. The 2 variants, mef(A) and mef(E), are 90% similar and were long regarded as a single gene class because they were not distinguishable by PCR [7, 8].
The genetic elements for the two variants are carried on different but related elements [7]. The mef(A) gene is carried on the defective transposon Tn1207.1 or the closely related Tn1207.3 and mef(E) on the "macrolide efflux genetic assembly" (mega) element. Other mobile genetic elements have also been described, such as the Tn916 family, which contains composite elements, such as tetM plus mega (Tn2009), and tetM, erm(B), and mega (Tn2010) [3].
Distribution of these transposons and the genes carried by them vary in different geographic regions [9], thereby providing an indication towards the origin of antibiotic-resistant strains of S. pneumoniae. It is, therefore, important to understand the distribution of macrolide resistance in different countries. In addition, analysis of the molecular genetics of erythromycin resistance by including the types of transposons and their resistance-conferring genes in clinical and normal flora pneumococci isolates with M phenotype would add to our present knowledge of the molecular epidemiology of S. pneumoniae.
Go to : 
The isolates were collected from patients admitted to the city hospitals and private laboratories in Tehran, Iran. The studies were carried out over a period of 24 months, from 2011 to 2013. Only one isolate per patient was included in the study. Nasopharynx normal flora isolates were collected from healthy people. Patients who had any antibiotic treatment in the last 6 months were excluded from the study. A consent form was filled by the participants.
Specimens were streaked on 5% sheep blood agar plate (Merck KGaA, Darmstadt, Germany) and were incubated for 24-48 hr at 37℃ in a candle jar. Standard microbiological techniques using typical colonial appearance, hemolysis, Gram staining, bile solubility, and susceptibility to optochin (1 µg) discs were performed for species identification. S. pneumonia ATCC 49619 was used for quality control. The final identification was confirmed by PCR using species-specific primers for amplification of lytA gene. The primer sets used were lytA-F: CGGACTAC GCCTTTATATCG and lytA-R: GTTTCAATCGTCAAGCCGTT [10].
Isolates were primarily identified as macrolide-lincosamide-streptograminB (MLSB)-resistant phenotypes by the double-disc diffusion method using erythromycin (15 µg) (Mast Diagnostics Ltd, Bootle, Merseyside, UK) and clindamycin (2 µg) (Mast Diagnostics Ltd) standards discs. Constitutive, inducible, or M phenotypes were determined by placing the clindamycin disc adjacent to erythromycin. After 24-hr incubation, flattened colonies around the clindamycin zone and in the area between the two discs (D zone) had an inducible MLSB (iMLSB) phenotype, whereas the absence of blunting indicated the presence of the constitutive MLSB (cMLSB) phenotype. The M phenotype was characterized by clindamycin susceptibility with no blunting of the inhibition zone around the clindamycin disc [11].
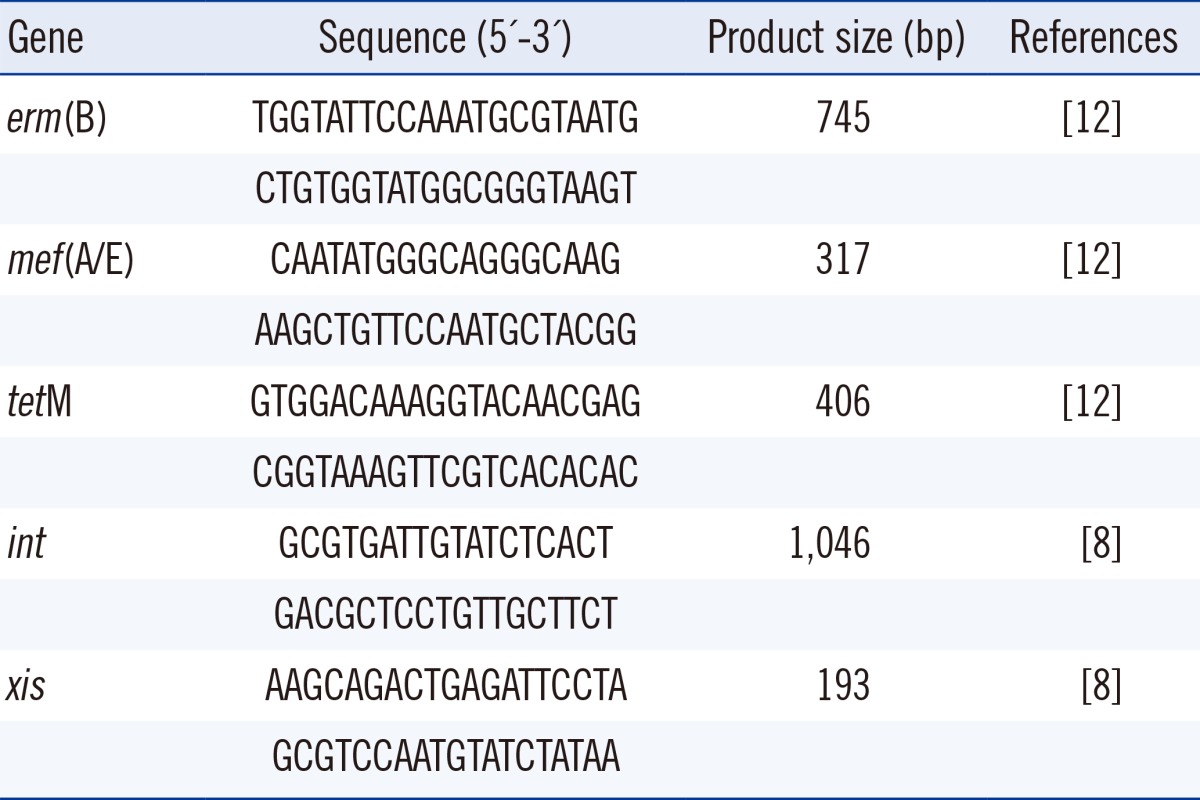
DNA extraction of pneumococcal isolates was done with peqGOLD Bacterial DNA Kit (peQlab, Erlangen, Germany). The erythromycin resistance and genetic elements genes were detected by PCR with two sets of primers specific for erm(B), mef(A/E), and tetM (Table 1) [12]. The PCR amplicons were produced by an initial cycle at 95℃ for 10 min, followed by 30 cycles of denaturation at 94℃ for 30 sec, annealing at 57℃ for 30 sec, and extension at 72℃ for 30 sec with a final extension at 72℃ for 5 min.
PCR-based confirmation for the presence of mef gene was performed with a set of primer pairs (Table 1) that also enabled recognition of the type of genetic organization of the transposon. The PCR program for int and xis genes were as follows: an initial cycle at 95℃ for 10 min, followed by 30 cycles of denaturation at 94℃ for 30 sec, annealing at 52℃ for 30 sec, and extension at 72℃ for 30 sec with a final extension at 72℃ for 5 min.
Isolates primarily identified as pneumococci with M phenotype were subsequently tested for resistance against tetracycline (30 µg), oxacillin (1 µg), clindamycin (2 µg), cotrimoxazole (1.25/23.75 µg), vancomycin (30 µg), and chloramphenicol (30 µg) (Mast Diagnostics Ltd) by employing the disc diffusion agar method.
Minimum inhibitory concentrations (MICs) of erythromycin were determined by the Etest (Liofilchem, Via Scozia, Italy) method on Mueller-Hinton agar with 5% defibrinated sheep blood according to the manufacturer's instructions. All plates were incubated at 37℃ for 20 hr. The MICs for penicillin for all isolates with the M phenotype were also determined by the Etest method. MIC results were interpreted according to the guidelines of the Clinical and Laboratory Standard Institute [13].
All pneumococcal isolates with the M phenotype were typed by using BOX-PCR with the boxA 5'-CTACGGCAAGGCGACGCTGACG-3'primer [14].
PCR for amplification was performed under the following conditions: pre-denaturation at 95℃ for 7 min, 30 cycles each of denaturation at 90℃ for 30 sec, primer annealing at 48℃ for 1 min, chain extension at 65℃ for 8 min, and a single cycle extension at 65℃ for 16 min. After amplification, products were elecrophoresed in a 1% agarose gel with 0.5×Tris-borate-EDTA at 100 V for 3 hr. The gel was stained with ethidium bromide (1 mg/mL) and viewed with UV light. Banding patterns that differed from each other by more than three bands were examined visually.
Go to : 
In our study, 92 clinical and 94 normal flora isolates were identified as pneumococci. Antibiotic susceptibility results for erythromycin revealed that 43 (46%) from clinical and 45 (49%) from normal flora isolates were resistant to erythromycin. Clinical isolates were obtained from blood (15), cerebrospinal fluid (6), lung secretions (9), eyes (8), sinuses (1), trachea (1), and ears (3).
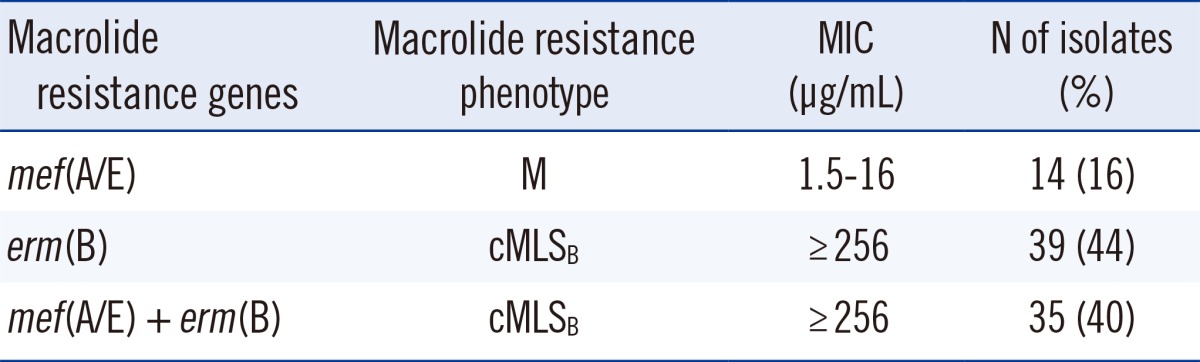
The double-disc diffusion test with erythromycin and clindamycin for the 88 test strains revealed that 74 (84%) indicated the cMLSB phenotype, and 14 (16%) were assigned to the M phenotype. These findings, together with the ranges of MICs of erythromycin, are summarized in Table 2. Amongst the isolates, erm(B) gene alone was detected in 39 (44%) and mef(A/E) in14 (16%) isolates, while 35 (40%) isolates harbored both erm(B) and mef(A/E) simultaneously as shown in Table 2. The association of tetM and mef gene was found in 100% of isolates with the M phenotype.
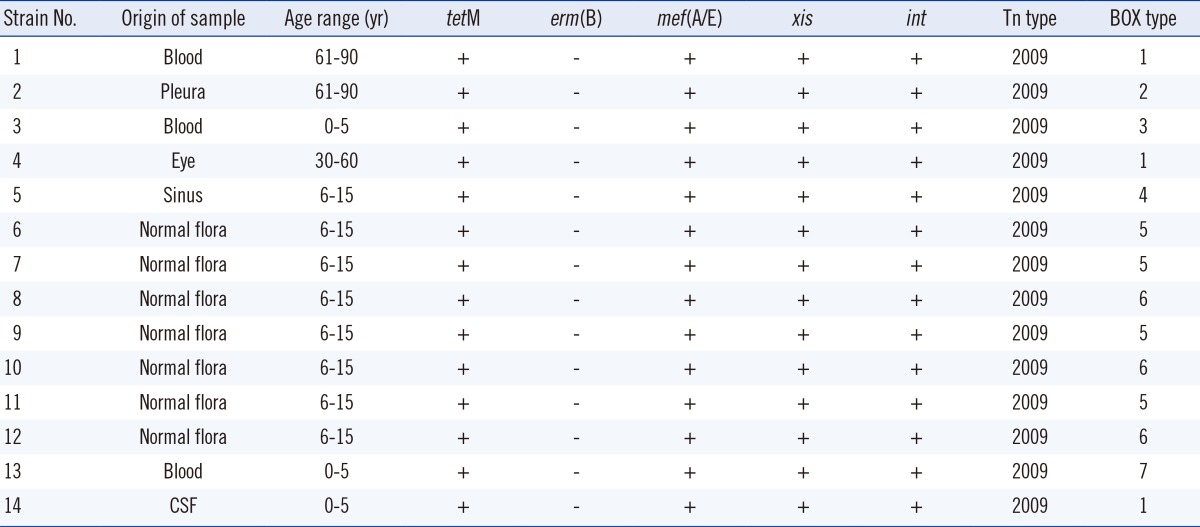
To gain further insights into the 14 strains with the M phenotype carrying the mef(A/E) genetic elements, we investigated the presence of two genes, xis and int, associated with Tn2009. The results revealed that all isolates carrying the mef gene were also positive for xis and int, thus exhibiting association with Tn2009 (Table 3).
All the resistant isolates with M phenotype were resistant to tetracycline (100%) and cotrimoxazole (100%), and 42% were also resistant to chloramphenicol, while they were all found to be susceptible to clindamycin and vancomycin. The erythromycin MIC for 84% of isolates with cMLSB phenotype was ≥256 µg/mL. The MIC for all of isolates with the M phenotype was 1.5-16 µg/mL (Table 2). Furthermore, results obtained from MIC for penicillin indicated only one resistant isolate (≥2 µg/mL), 3 isolates with high susceptibility (<0.1 µg/mL), and the remaining within intermediate susceptibility (0.1-1.5 µg/mL).
According to the BOX PCR typing method, the erythromycin-resistant strains with M phenotype belonged to 3 common types with 10 isolates and 4 single types (Table 3). All normal flora samples were divided in two common types, whilst the clinical samples included one common type and four single types.
Go to : 
There has been an increase in the numbers of erythromycin-resistant S. pneumoniae strains worldwide [3]. The previous year also witnessed a worldwide increase in the prevalence of efflux mechanisms (M phenotype) amongst the S. pneumoniae strains [15]. In line with this, our results show a high prevalence of resistance to macrolides (47%), and the M phenotype was found in 16% of the isolates, all of which carried mef(A/E). These values are much higher than that determined in other studies, 9.1% in Spain and 3.1% in France [9, 16] reporting lower prevalence of mef(A/E) among macrolide-resistant isolates. Our results are also consistent with the studies on pneumococcal macrolide resistance, which show that it is more prevalent in the United States and Canada, with a higher rate of M phenotype and mef(A/E) genotype compared with European countries [16].
Almost 40% of the pneumococcal isolates obtained from Korea and South Africa have been reported to carry erm(B) and mef(A/E) genes [17, 18]. Moreover, other countries have also reported, to lesser extent, simultaneous presence of the two genes [3, 7, 18, 19]. Overall, all the reports suggest an emergence of resistant pneumococci erm(B) and mef(E) genes worldwide.
As expected, the present results showed that the isolates with erm(B) gene alone or with mef were highly resistant to erythromycin. On the other hand, the strains with mef(A/E) alone showed low-level resistance to erythromycin, which is the typical characteristic of strains with the M phenotype [18, 20].
Here, we showed that the isolates with M phenotype were associated with high rates of chloramphenicol, cotrimoxazole, and tetracycline resistance. In addition, penicillin remains a therapeutic choice for all of the isolates (10 intermediate and 1 resistant), which were detected as susceptible, except for three penicillin-resistant isolates. The results obtained are consistent with other reports suggesting concomitant resistance to erythromycin and other antibiotics with therapeutic options [20, 21]. Furthermore, we observed that all isolates with M phenotype were also resistant to tetracycline. The resistance to tetracycline has frequently been associated with erythromycin [7].
The high resistance rate to tetracycline might also be due to the presence of Tn2009 encoding genes conferring tetracycline and erythromycin resistance. All of the isolates with mef(A/E) were positive for int and xis by PCR. This combination of genes is characteristic of Tn2009, which is a Tn916-related transposon. Cochetti et al. [8] have also reported the mega element as a major element carrying mef(A/E) gene. This transposon was detected in other studies as the genetic element conferring macrolide resistance in pneumococcal isolates [9, 22].
Using BOX PCR typing, we showed the pneumococci with M phenotype in clinical isolates to be polyclonal, whereas normal flora isolates were clonally related. As reported earlier [16], the emergence of efflux mechanism of macrolide resistance is not clonally related in clinical isolates. They can, however, be transferred clonally.
In conclusion, the rate of macrolide resistance in our S. pneumoniae collection was high. Although the cMLSB phenotype predominates macrolide resistance in Iran, the M phenotype, appears to be emerging as an important factor in erythromycin-resistant pneumococci. A majority of these isolates showed a high level of resistance to tetracycline, cotrimoxazole, and penicillin, which makes treatment difficult. It is, therefore, necessary to collect data from various countries for the worldwide surveillance on emergence of macrolide resistance in S. pneumoniae isolates.
Go to : 
Acknowledgments
The work was financially supported by Iran University of Medical Sciences with grant number 21674.
Go to : 
References
1. Henderson KL, Muller-Pebody B, Blackburn RM, Johnson AP. Reduction in erythromycin resistance in invasive pneumococci from young children in England and Wales. J Antimicrob Chemother. 2010; 65:369–370. PMID: 20007730.

2. J J, M T N, M D MN, M SN, M Y R, J F, et al. Prevalence of macrolide resistance and in vitro activities of six antimicrobial agents against clinical isolates of Streptococcus pneumoniae from a multi-center surveillance in Malaysia. Med J Malaysia. 2013; 68:119–124. PMID: 23629556.
3. Xu X, Cai L, Xiao M, Kong F, Oftadeh S, Zhou F, et al. Distribution of serotypes, genotypes, and resistance determinants among macrolide-resistant Streptococcus pneumoniae isolates. Antimicrob Agents Chemother. 2010; 54:1152–1159. PMID: 20065057.
4. Pihlajamäki M, Kataja J, Seppälä H, Elliot J, Leinonen M, Huovinen P, et al. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob Agents Chemother. 2002; 46:654–658. PMID: 11850244.
5. Taha N, Araj GF, Wakim RH, Kanj SS, Kanafani ZA, Sabra A, et al. Genotypes and serotype distribution of macrolide resistant invasive and non-invasive Streptococcus pneumoniae isolates from Lebanon. Ann Clin Microbiol Antimicrob. 2012; 11:2. PMID: 22248318.

6. Sangvik M, Littauer P, Simonsen GS, Sundsfjord A, Dahl KH. mef(A), mef(E) and a new mef allele in macrolide-resistant Streptococcus spp. isolates from Norway. J Antimicrob Chemother. 2005; 56:841–846. PMID: 16172106.
7. Montanari MP, Cochetti I, Mingoia M, Varaldo PE. Phenotypic and molecular characterization of tetracycline- and erythromycin-resistant strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2003; 47:2236–2241. PMID: 12821474.
8. Cochetti I, Vecchi M, Mingoia M, Tili E, Catania MR, Manzin A, et al. Molecular characterization of pneumococci with efflux-mediated erythromycin resistance and identification of a novel mef gene subclass, mef(I). Antimicrob Agents Chemother. 2005; 49:4999–5006. PMID: 16304164.
9. Calatayud L, Ardanuy C, Tubau F, Rolo D, Grau I, Pallarés R, et al. Serotype and genotype replacement among macrolide-resistant invasive Pneumococci in adults: mechanisms of resistance and association with different transposons. J Clin Microbiol. 2010; 48:1310–1316. PMID: 20147647.

10. Strålin K, Korsgaard J, Olcén P. Evaluation a multiplex PCR for bacterial pathogens applied to bronchoalveolar lavage. Eur Respir J. 2006; 28:568–575. PMID: 16737990.
11. Zhou L, Ma X, Gao W, Yao KH, Shen AD, Yu SJ, et al. Molecular characteristics of erythromycin-resistant Streptococcus pneumoniae from pediatric patients younger than five years in Beijing, 2010. BMC Microbiol. 2012; 12:228–238. PMID: 23043378.

12. Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005; 49:4798–4800. PMID: 16251336.

13. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 23rd Informational supplement, M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute;2013.
14. Payne as DB, Sun A, Butler JC, Singh SP, Hollingshead SK, Briles DE. PspA family typing and PCR-based DNA fingerprinting with BOX A1R primer of pneumococci from the blood of patients in the USA with and without sickle cell disease. Epidemiol Infect. 2005; 133:173–178. PMID: 15724724.
15. de la Pedrosa EG, Morosini MI, van der Linden M, Ruiz-Garbajosa P, Galán JC, Baquero F, et al. Polyclonal population structure of Streptococcus pneumoniae isolates in Spain carrying mef and mef plus erm(B). Antimicrob Agents Chemother. 2008; 52:1964–1969. PMID: 18362188.
16. Marchandin H, Jean-Pierre H, Jumas-Bilak E, Isson L, Drouillard B, Darbas H, et al. Distribution of macrolide resistance genes erm(B) and mef(A) among 160 penicillin-intermediate clinical isolates of Streptococcus pneumoniae isolated in southern France. Pathol Biol (Paris). 2001; 49:522–527. PMID: 11642013.
17. Bae S, Lee K. Distribution of capsular serotypes and macrolide resistance mechanisms among macrolide-resistant Streptococcus pneumoniae isolates in Korea. Diagn Microbiol Infect Dis. 2009; 63:213–216. PMID: 19097840.
18. McGee L, Klugman KP, Wasas A, Capper T, Brink A. Serotype 19f multiresistant pneumococcal clone harboring two erythromycin resistance determinants (erm(B) and mef(A)) in South Africa. Antimicrob Agents Chemother. 2001; 45:1595–1598. PMID: 11302838.
19. Cochetti I, Tili E, Vecchi M, Manzin A, Mingoia M, Varaldo PE, et al. New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J Antimicrob Chemother. 2007; 60:127–131. PMID: 17483548.
20. Felmingham D, Cantón R, Jenkins SG. Regional trends in beta-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J Infect. 2007; 55:111–118. PMID: 17568680.
21. Hsueh PR, Teng LJ, Lee LN, Yang PC, Ho SW, Luh KT. Extremely high incidence of macrolide and trimethoprim-sulfamethoxazole resistance among clinical isolates of Streptococcus pneumoniae in Taiwan. J Clin Microbiol. 1999; 37:897–901. PMID: 10074498.
22. Calatayud L, Ardanuy C, Cercenado E, Fenoll A, Bouza E, Pallares R, et al. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob Agents Chemother. 2007; 51:3240–3246. PMID: 17606677.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download