Abstract
Carbapenemase production has been reported worldwide in gram-negative bacteria, including Acinetobacter species. We detected carbapenemase-producing Acinetobacter pittii in clinical isolates in Daejeon, Korea. Twenty-one ertapenem-resistant A. pittii isolates screened with a disk diffusion method were characterized by using the Epsilon test, four multiplex PCR assays, and a multilocus sequence typing (MLST) scheme. A total of 21 A. pittii isolates harbored the metallo-β-lactamase (MBL) gene blaIMP-1 or blaNDM-1. Nineteen isolates containing blaIMP-1 were resistant to imipenem and meropenem, but two isolates harboring blaNDM-1 were susceptible to them. The sequence types (STs) of the two New Delhi MBL (NDM-1)-producing A. pittii isolates were ST70 and ST207, which differed from the STs (ST63, ST119, ST396, and a novel ST) of the IMP-1-producing A. pittii. This is the first report on NDM-1-producing A. pittii isolates in Korea. Our results emphasize that the study of NDM-1-producing gram-negative bacteria should involve carbapenem-susceptible as well as carbapenem-resistant isolates.
Acinetobacter species are important nosocomial and opportunistic pathogens that cause frequent outbreaks in intensive care units. Most strains involved in nosocomial infections are highly resistant to various antimicrobial agents and cause life-threatening illnesses. Carbapenems have been the drugs of choice to treat Acinetobacter infection because they are stable in response to extended-spectrum and AmpC β-lactamases. However, carbapenem-resistant Acinetobacter strains have been increasingly reported [12].
Carbapenemase production is the most critical mechanism of carbapenem resistance, and various carbapenemases, such as the Ambler class B metallo-β-lactamases (MBLs) of IMP, GIM, SIM, SPM, and VIM types and the Ambler class D carbapenemases of OXA-23, OXA-40, OXA-58, and OXA-143 types, have been reported in Acinetobacter species worldwide [3]. In addition, New Delhi MBL (NDM-1) has recently emerged as a key carbapenemase that compromises the efficiency of almost all β-lactams.
Although carbapenemase genes are widely disseminated among Acinetobacter species isolates, few data are available for Acinetobacter pittii isolates harboring carbapenemase. Clinically, Acinetobacter baumannii is the most commonly isolated species, but A. pittii isolates are also detected frequently and show resistance to various antimicrobial agents. In this study, we investigated carbapenemase genes in A. pittii isolates from a university hospital in Daejeon, Korea. In addition, multilocus sequence typing (MLST) was performed to analyze the relationship between carbapenemase types and A. pittii isolates harboring carbapenemases.
A. pittii isolates were collected at a university hospital laboratory in Daejeon, Korea, between January 2006 and December 2013. A. pittii was identified by using the Vitek 2 Automated Instrument ID System (bioMérieux; Marcy l'Etoile, France) and by sequencing the partial rpoB housekeeping gene as described previously [4].
To investigate the presence of carbapenemases, we performed disk diffusion tests using 10-µg ertapenem disks (BD, Franklin Lakes, NJ, USA). All isolates with inhibition zone diameters of ≤21 mm on the ertapenem disks were subjected to carbapenemase detection. The minimum inhibitory concentrations (MICs) of A. pittii isolates for imipenem, meropenem, ceftazidime, and cefepime were determined with the Epsilon test (Etest; bioMérieux). The data were interpreted according to the criteria approved by CLSI [5]. Escherichia coli ATCC 25922 was used as a reference strain.
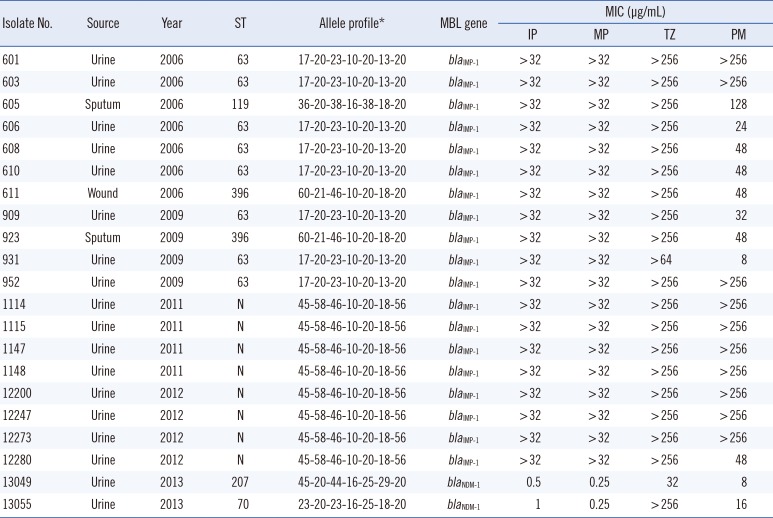
A total of 21 ertapenem-resistant A. pittii isolates were recovered from urine, sputum, or wound specimens. Most of these isolates (85.7%) were obtained from urine specimens (Table 1). The isolates were subjected to four multiplex PCR assays for the detection of carbapenemase genes. These assays were defined as multi-1, multi-2, multi-3, and multi-4 and were composed of specific primer sets [678]. Total DNA was obtained from each target strain by using a DNA purification kit (SolGent, Daejeon, Korea) in accordance with standard protocols. Each multiplex PCR was performed in a total volume of 25 µL with 50 ng total DNA, 2.5 µL 10× Taq buffer, 0.5 µL 10 mM dNTP mix, 20 pmol each primer, and 0.7 U Taq DNA polymerase (SolGent). PCR was performed in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer Cetus Corp., Norwalk, CT, USA) with pre-denaturation of the reaction mixture at 95℃ for 5 min, followed by 35 cycles at 95℃ for 30 sec, 52℃ for 40 sec, and 72℃ for 30 sec, with a final extension at 72℃ for 5 min. The amplified products were separated via electrophoresis on 1.5% (w/v) agarose gels containing ethidium bromide and visualized with a BioDoc-14TM Imaging system (UVP, Cambridge, UK).
All positive isolates from the multiplex PCR were subjected to PCR assays with specific primers to identify carbapenemase genes [9]. NDM-1-positive A. pittii isolates were also subjected to PCR with previously described primers to analyze the surrounding structure of blaNDM-1 [10]. The amplicons were purified with a PCR purification kit (SolGent) and sequenced using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3730XL DNA analyzer (PE Applied Biosystems). The various DNA sequences were confirmed with the Basic Local Alignment Search Tool (BLAST) paired alignment facility (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
An MLST scheme with seven housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) was used to determine the sequence types (STs) of A. pittii isolates [11]. Each ST number was assigned by comparing the allele sequences to those in the MLST databases (http://www.pasteur.fr/mlst and http://pubmlst.org/abaumannii).
All 21 consecutive A. pittii isolates resistant to ertapenem contained blaIMP-1 or blaNDM-1 (Table 1). However, other MBL genes and OXA-type carbapenemases were not found within the cohort.
Most of the ertapenem-resistant A. pittii isolates (90.5%) harbored blaIMP-1, and all of these isolates showed high resistance to imipenem (MICs ≥32 µg/mL) and meropenem (MICs ≥32 µg/mL). The frequency of these isolates was in accordance with that of previously reported blaIMP-1-containing Acinetobacter species isolated in Korea [12]. Our results indicate that blaIMP-1 is widely disseminated among ertapenem-resistant A. pittii isolates at the university hospital in Daejeon, Korea.
Interestingly, two ertapenem-resistant A. pittii isolates in this study contained blaNDM-1, which has been detected in Enterobacteriaceae clinical isolates in South Korea [13]. This study is the first to report an NDM-1-type MBL in A. pittii isolated in Korea. Because NDM-1 was first described in carbapenem-resistant Klebsiella pneumoniae and Escherichia coli isolates, this MBL has now been found in various Enterobacteriaceae, Pseudomonas, and Acinetobacter species [14]. Although NDM-1-producing isolates are usually resistant to carbapenems, the isolates harboring blaNDM-1 in our study were susceptible to imipenem and meropenem [10]. This result is similar to that of previous reports that some NDM-1 producers among Enterobacteriaceae and Acinetobacter showed lower MIC values to imipenem and meropenem [1516]. Therefore, ertapenem rather than other carbapenems has been proposed for the detection of NDM producers [17].
Consequently, we analyzed the genetic environment of blaNDM-1 in the two A. pittii isolates and confirmed an approximately 3,500-bp nucleotide segment containing aminoglycoside resistance gene aphA6 and transposase ISAba125 upstream and a truncated bleomycin resistance gene and trpF genes downstream of blaNDM-1. The segment detected in our study was identical to that reported in the previous study of Acinetobacter species [10], which reported that ISAba125 upstream of blaNDM-1 may be the main explanation for blaNDM-1 gene dissemination.
In this study, we performed MLST for epidemiological typing of ertapenem-resistant A. pittii isolates. The two bacterial isolates containing blaNDM-1 belonged to ST70 and ST207, which were different from the STs of the isolates harboring blaIMP-1 (ST63, ST119, ST396, and a novel ST). In previous studies, blaNDM-1 was identified in A. pittii isolates of ST63 in China [18], and NDM-1-producing strains isolated in Paraguay displayed ST320 and ST321 [19]. MLST profiles for four isolates containing blaNDM-1 identified in China differed from those described previously [10]. In addition, A. pittii isolates with ST63 and ST119 in Japan harbored blaIMP-11 and blaIMP-19, respectively [20]. Although the databases of MLST profiles for A. pittii are poor, our results suggest that ST was not directly related to MBL production in A. pittii isolates. Furthermore, the two strains harboring blaNDM-1 were all isolated in 2013, whereas the other strains were isolated before 2012. Our results suggest that A. pittii isolates harboring blaNDM-1 emerged only recently in Korea.
The emergence and dissemination of carbapenemase genes, including the NDM gene, in Acinetobacter species has become a major concern in the treatment of bacterial infections with antimicrobial agents. Early recognition and detection of carbapenemases are critical in controlling infection by clinical Acinetobacter isolates showing high resistance to carbapenems. On the basis of the findings that some carbapenemase producers can show susceptible phenotypes and that the NDM gene has a high propensity to disseminate, we suggest that the detection of carbapenem-producing isolates should extend to clinical isolates of carbapenem-susceptible Acinetobacter species in addition to carbapenem-resistant ones. In particular, additional tests are needed to detect carbapenemase producers with carbapenem susceptibility.
References
1. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006; 12:826–836. PMID: 16882287.
2. Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007; 2:501–512. PMID: 17927473.

3. Nordmann P. Gram-negative bacteriae with resistance to carbapenems. Med Sci (Paris). 2010; 26:950–959. PMID: 21106177.
4. Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009; 155:2333–2341. PMID: 19389786.
5. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement, M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute;2010.
6. Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007; 59:321–322. PMID: 17185300.
7. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011; 70:119–123. PMID: 21398074.

8. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006; 27:351–353. PMID: 16564159.
9. Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrug-resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010; 30:498–506. PMID: 20890082.
10. Zhang R, Hu YY, Yang XF, Gu DX, Zhou HW, Hu QF, et al. Emergence of NDM-producing non-baumannii Acinetobacter spp. isolated from China. Eur J Clin Microbiol Infect Dis. 2014; 33:853–860. PMID: 24306098.
11. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010; 5:e10034. PMID: 20383326.
12. Lee K, Kim MN, Choi TY, Cho SE, Lee S, Whang DH, et al. Wide dissemination of OXA-type carbapenemases in clinical Acinetobacter spp. isolates from South Korea. Int J Antimicrob Agents. 2009; 33:520–524. PMID: 19091520.
13. Yoo JS, Kim HM, Koo HS, Yang JW, Yoo JI, Kim HS, et al. Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J Antimicrob Chemother. 2013; 68:2170–2172. PMID: 23696618.
14. Janvier F, Jeannot K, Tessé S, Robert-Nicoud M, Delacour H, Rapp C, et al. Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob Agents Chemother. 2013; 57:3408–3411. PMID: 23612200.
15. Zong Z, Zhang X. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother. 2013; 68:1007–1010. PMID: 23288403.
16. Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011; 49:718–721. PMID: 21123531.

17. Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int. 2014; 2014:249856. PMID: 24790993.

18. Yang J, Chen Y, Jia X, Luo Y, Song Q, Zhao W, et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect. 2012; 18:E506–E513. PMID: 23036089.
19. Pasteran F, Mora MM, Albornoz E, Faccone D, Franco R, Ortellado J, et al. Emergence of genetically unrelated NDM-1-producing Acinetobacter pittii strains in Paraguay. J Antimicrob Chemother. 2014; 69:2575–2578. PMID: 24793901.
20. Yamamoto M, Nagao M, Matsumura Y, Matsushima A, Ito Y, Takakura S, et al. Interspecies dissemination of a novel class 1 integron carrying blaIMP-19 among Acinetobacter species in Japan. J Antimicrob Chemother. 2011; 66:2480–2483. PMID: 21862476.
Table 1
Characteristics of Acinetobacter pittii isolates harboring blaMBL genes




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download