INTRODUCTION
METHODS
1. Patients
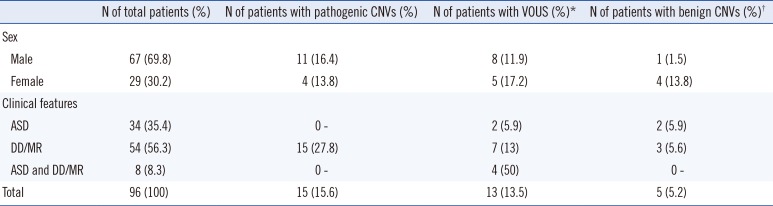
Table 1
Number of patients with copy number variations according to demographic and clinical features
2. DNA preparation
3. MLPA
4. Chromosomal microarray
5. Interpretation of detected CNVs
RESULTS
1. Results of MLPA and CMA
Table 2
Summary of clinical features and genetic test results of patients with copy number variants or loss of heterozygosity
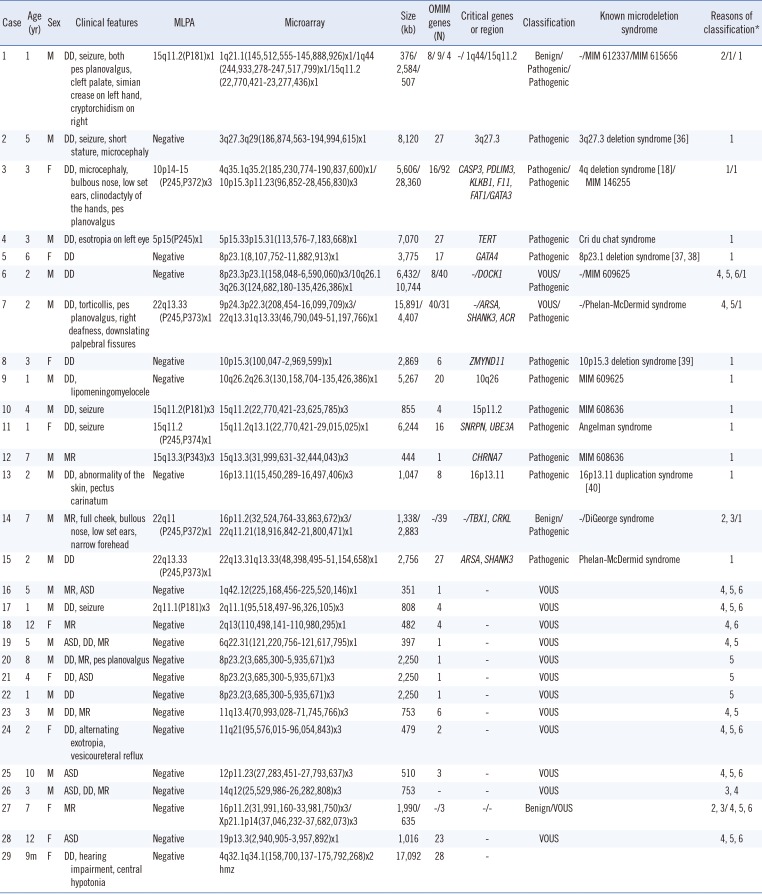
| Case | Age (yr) | Sex | Clinical features | MLPA | Microarray | Size (kb) | OMIM genes (N) | Critical genes or region | Classification | Known microdeletion syndrome | Reasons of classification* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | M | DD, seizure, both pes planovalgus, cleft palate, simian crease on left hand, cryptorchidism on right | 15q11.2(P181)x1 | 1q21.1(145,512,555-145,888,926)x1/1q44 | 376/2,584/507 | 8/ 9/ 4 | -/ 1q44/15q11.2 | Benign/Pathogenic/Pathogenic | -/MIM 612337/MIM 615656 | 2/1/ 1 |
| (244,933,278-247,517,799)x1/15q11.2 | |||||||||||
| (22,770,421-23,277,436)x1 | |||||||||||
| 2 | 5 | M | DD, seizure, short stature, microcephaly | Negative | 3q27.3q29(186,874,563-194,994,615)x1 | 8,120 | 27 | 3q27.3 | Pathogenic | 3q27.3 deletion syndrome [36] | 1 |
| 3 | 3 | F | DD, microcephaly, bulbous nose, low set ears, clinodactyly of the hands, pes planovalgus | 10p14-15 (P245,P372)x3 | 4q35.1q35.2(185,230,774-190,837,600)x1/ | 5,606/28,360 | 16/92 | CASP3, PDLIM3, KLKB1, F11, FAT1/GATA3 | Pathogenic/Pathogenic | 4q deletion syndrome [18]/MIM 146255 | 1/1 |
| 10p15.3p11.23(96,852-28,456,830)x3 | |||||||||||
| 4 | 3 | M | DD, esotropia on left eye | 5p15(P245)x1 | 5p15.33p15.31(113,576-7,183,668)x1 | 7,070 | 27 | TERT | Pathogenic | Cri du chat syndrome | 1 |
| 5 | 6 | F | DD | Negative | 8p23.1(8,107,752-11,882,913)x1 | 3,775 | 17 | GATA4 | Pathogenic | 8p23.1 deletion syndrome [3738] | 1 |
| 6 | 2 | M | DD | Negative | 8p23.3p23.1(158,048-6,590,060)x3/10q26.1 | 6,432/10,744 | 8/40 | -/DOCK1 | VOUS/Pathogenic | -/MIM 609625 | 4, 5, 6/1 |
| 3q26.3(124,682,180-135,426,386)x1 | |||||||||||
| 7 | 2 | M | DD, torticollis, pes planovalgus, right deafness, downslating palpebral fissures | 22q13.33 (P245,P373)x1 | 9p24.3p22.3(208,454-16,099,709)x3/ | 15,891/4,407 | 40/31 | -/ARSA, SHANK3, ACR | VOUS/Pathogenic | -/Phelan-McDermid syndrome | 4, 5/1 |
| 22q13.31q13.33(46,790,049-51,197,766)x1 | |||||||||||
| 8 | 3 | F | DD | Negative | 10p15.3(100,047-2,969,599)x1 | 2,869 | 6 | ZMYND11 | Pathogenic | 10p15.3 deletion syndrome [39] | 1 |
| 9 | 1 | M | DD, lipomeningomyelocele | Negative | 10q26.2q26.3(130,158,704-135,426,386)x1 | 5,267 | 20 | 10q26 | Pathogenic | MIM 609625 | 1 |
| 10 | 4 | M | DD, seizure | 15q11.2(P181)x3 | 15q11.2(22,770,421-23,625,785)x3 | 855 | 4 | 15p11.2 | Pathogenic | MIM 608636 | 1 |
| 11 | 1 | F | DD, seizure | 15q11.2(P245,P374)x1 | 15q11.2q13.1(22,770,421-29,015,025)x1 | 6,244 | 16 | SNRPN, UBE3A | Pathogenic | Angelman syndrome | 1 |
| 12 | 7 | M | MR | 15q13.3(P343)x3 | 15q13.3(31,999,631-32,444,043)x3 | 444 | 1 | CHRNA7 | Pathogenic | MIM 608636 | 1 |
| 13 | 2 | M | DD, abnormality of the skin, pectus carinatum | Negative | 16p13.11(15,450,289-16,497,406)x3 | 1,047 | 8 | 16p13.11 | Pathogenic | 16p13.11 duplication syndrome [40] | 1 |
| 14 | 7 | M | MR, full cheek, bullous nose, low set ears, narrow forehead | 22q11(P245,P372)x1 | 16p11.2(32,524,764-33,863,672)x3/ | 1,338/2,883 | -/39 | -/TBX1, CRKL | Benign/Pathogenic | -/DiGeorge syndrome | 2, 3/1 |
| 22q11.21(18,916,842-21,800,471)x1 | |||||||||||
| 15 | 2 | M | DD | 22q13.33(P245,P373)x1 | 22q13.31q13.33(48,398,495-51,154,658)x1 | 2,756 | 27 | ARSA, SHANK3 | Pathogenic | Phelan-McDermid syndrome | 1 |
| 16 | 5 | M | MR, ASD | Negative | 1q42.12(225,168,456-225,520,146)x1 | 351 | 1 | - | VOUS | 4, 5, 6 | |
| 17 | 1 | M | DD, seizure | 2q11.1(P181)x3 | 2q11.1(95,518,497-96,326,105)x3 | 808 | 4 | VOUS | 4, 5, 6 | ||
| 18 | 12 | F | MR | Negative | 2q13(110,498,141-110,980,295)x1 | 482 | 4 | - | VOUS | 4, 6 | |
| 19 | 5 | M | ASD, DD, MR | Negative | 6q22.31(121,220,756-121,617,795)x1 | 397 | 1 | - | VOUS | 4, 5 | |
| 20 | 8 | M | DD, MR, pes planovalgus | Negative | 8p23.2(3,685,300-5,935,671)x3 | 2,250 | 1 | - | VOUS | 5 | |
| 21 | 4 | F | DD, ASD | Negative | 8p23.2(3,685,300-5,935,671)x3 | 2,250 | 1 | - | VOUS | 5 | |
| 22 | 1 | M | DD | Negative | 8p23.2(3,685,300-5,935,671)x3 | 2,250 | 1 | - | VOUS | 5 | |
| 23 | 3 | M | DD, MR | Negative | 11q13.4(70,993,028-71,745,766)x3 | 753 | 6 | - | VOUS | 4, 5 | |
| 24 | 2 | F | DD, alternating exotropia, vesicoureteral reflux | Negative | 11q21(95,576,015-96,054,843)x3 | 479 | 2 | - | VOUS | 4, 5, 6 | |
| 25 | 10 | M | ASD | Negative | 12p11.23(27,283,451-27,793,637)x3 | 510 | 3 | - | VOUS | 4, 5, 6 | |
| 26 | 3 | M | ASD, DD, MR | Negative | 14q12(25,529,986-26,282,808)x3 | 753 | - | - | VOUS | 3, 4 | |
| 27 | 7 | F | MR | Negative | 16p11.2(31,991,160-33,981,750)x3/ | 1,990/635 | -/3 | -/- | Benign/VOUS | 2, 3/ 4, 5, 6 | |
| Xp21.1p14(37,046,232-37,682,073)x3 | |||||||||||
| 28 | 12 | F | ASD | Negative | 19p13.3(2,940,905-3,957,892)x1 | 1,016 | 23 | - | VOUS | 4, 5, 6 | |
| 29 | 9m | F | DD, hearing impairment, central hypotonia | Negative | 4q32.1q34.1(158,700,137-175,792,268)x2 | 17,092 | 28 | - | |||
| hmz |
*1.Overlapping with a known genomic imbalance syndrome; 2. In the category of genomic imbalance in healthy individuals as per public database; 3. No OMIM genes in the CNV interval; 4. CNV is not a common polymorphism; 5. OMIM genes in the CNV interval are not known for dosage sensitivity; 6. OMIM genes in the CNV interval are not associated with patient's phenotype.
Abbreviations: MLPA, multiplex ligation-dependent probe amplification; VOUS, variant of uncertain clinical significance; ASD, autism spectrum disorder; DD, developmental delay; MR, mental retardation; m, months.
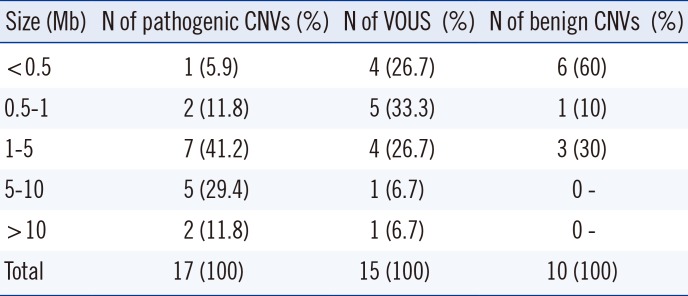
Table 3
Size distribution of pathogenic CNVs found in patients




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download