Abstract
Background
Peptide nucleic acid (PNA) probes are artificial DNA analogues with a hydrophobic nature that can penetrate the mycobacterial cell wall. We evaluated a FISH method for simultaneous detection and identification of Mycobacterium tuberculosis (MTB) and non-tuberculous mycobacteria (NTM) in clinical respiratory specimens using differentially labeled PNA probes.
Methods
PNA probes targeting the mycobacterial 16S ribosomal RNA were synthesized. The cross-reactivity of MTB- and NTM-specific probes was examined with reference strains and 10 other frequently isolated bacterial species. A total of 140 sputum specimens were analyzed, comprising 100 MTB-positive specimens, 21 NTM-positive specimens, and 19 MTB/NTM-negative specimens; all of them were previously confirmed by PCR and culture. The PNA FISH test results were graded by using the United States Centers for Disease Control and Prevention-recommended scale and compared with the results from the fluorochrome acid-fast bacterial stain.
Results
The MTB- and NTM-specific PNA probes showed no cross-reactivity with other tested bacterial species. The test results demonstrated 82.9% agreement with the culture results with diagnostic sensitivity of 80.2% and diagnostic specificity of 100.0% (kappa=0.52, 95% confidence interval: 0.370-0.676).
Conclusions
Dual-color PNA FISH showed high specificity for detecting and identifying mycobacteria in clinical specimens. However, because of its relatively low sensitivity, this method could be more applicable to culture confirmation. In application to direct specimens, the possibility of false-negative results needs to be considered.
Mycobacterial pathogenicity in humans differs from one mycobacterial species to another. Mycobacterium tuberculosis (MTB) and related species can cause tuberculosis (TB) in humans. TB remains one of the world's deadliest communicable diseases. In 2013, an estimated 9.0 million people developed TB, and 1.5 million died from the disease [1]. Numerous other mycobacterial species, often termed non-tuberculous mycobacteria (NTM), are present in the environment and are generally less pathogenic than MTB. NTM usually cause disease in patients who are immunocompromised.
When mycobacterial infection is suspected, rapid and accurate identification of the species is necessary. Rapid identification of the MTB complex can avoid unnecessary isolation of patients infected with NTM and implementation of toxic treatment regimens [2].
Mycobacterial infection is typically identified by microscopic examination of a sputum smear, growth characteristics, and biochemical and molecular tests. Conventional microscopic examination is simple, fast, inexpensive, and highly specific in areas where there is a high prevalence of TB. Thus, it is an important screening tool. However, microscopic examination has low diagnostic sensitivity because of the necessity for large numbers of microorganisms to produce a positive result, and yields poor positive predictive values for MTB [3]. Also, it does not differentiate MTB from NTM.
The current gold standard for TB diagnosis is culture. Culture is about 500 times more sensitive than microscopy and permits further investigations, including identification. However, culture techniques routinely take 6-9 weeks for a diagnosis to be confirmed. Therefore, several amplification-based techniques have been developed to speed detection and increase the sensitivity of TB identification.
Several studies have used peptide nucleic acid (PNA) probes for the identification of bacteria from various specimens [456]. PNA probes are artificially synthesized DNA analogues with an uncharged peptide backbone, which have more favorable hybridization properties and chemical, thermal, and biological stability than previously used probes. The absence of a charge and the high conformational flexibility of PNA probes allow them to hybridize with high specificity to complimentary DNA or RNA [7]. The hydrophobic structure of PNA probes gives improved penetration through the mycobacterial cell wall, yielding increased sensitivity [6].
We developed a FISH method using differentially labeled PNA probes for simultaneous identification and differentiation of MTB and NTM on a single slide, and evaluated its diagnostic applicability in clinical specimens.
Fluorescence-labeled PNA probes targeting the MTB or NTM 16S ribosomal RNA (rRNA) were synthesized by PANAGENE (Daejeon, Korea). The probe sequences were subsequently matched with sequences in the GenBank and EMBL databases by using BLAST analysis [8]. The probes specific for MTB or NTM were as follows: MTB probe, NH2-CTTAGGAATTTTCGG-Lys-Flu-H; NTM, NH2-CGGTCGCCCATTACG-Ala-Ala-Flu-H (where Flu is the fluorescent label) [6]. Probes were purified by reverse-phase HPLC at 50℃ and characterized by using an AXIMA-CFR matrix-assisted laser decomposition/ionization time-of-flight mass spectrometry instrument (Shimadzu Biotech, Columbia, MD, USA). The N-terminus of the NTM PNA probes was labeled with cyanine 3 fluorescent dye (Amersham, Piscataway, NJ, USA) and that of MTB PNA probes was labeled with 6-carboxyfluorescein fluorescent dye (MWG, Ebersberg, Germany).
To check cross-reactivity, we used two standard reference strains (MTB ATCC 13950 and M. kansasii ATCC 12479), clinical isolates of mycobacteria (M. abscessus, M. avium, and M. intracellulare), and 10 frequently isolated respiratory bacteria (Enterococcus faecalis, E. faecium, Staphylococcus aureus, Streptococcus pneumoniae, Nocardia asteroides, Escherichia coli, Haemophilus influenzae, Klebsiella pneumoniae, Proteus mirabilis, and Pseudomonas aeruginosa) (Table 1). A mixture of M. tuberculosis ATCC 13950 and M. kansasii ATCC 12479 was also used.
Bacterial suspensions were prepared as follows. Mycobacteria were grown on 3% Ogawa solid medium (Asan Pharmaceutical, Seoul, Korea) and in BacT/ALERT MP liquid medium (BioMérieux, Durham, NC, USA) at 37℃ until visible colonies or positive growth signals were observed. After solid-medium culture, a loop of colonies was suspended in 500 µL of phosphate-buffered saline (PBS). After liquid-medium culture, culture bottles were vortexed for 10-15 sec, and 1 mL of the culture was centrifuged for 2 min at 12,000g. The supernatant was removed, and the sediment was resuspended in 500 µL of PBS. For non-mycobacterial species, all strains were grown on blood agar plates at 37℃ in a 5% CO2 incubator for 24 hr. A loop of colonies was suspended in 500 µL of PBS. Twenty microliters of each suspension was added to microscope slides. Each smear was air dried and heated at 80℃ for 2 hr.
A total of 140 clinical sputum specimens during the period from May to October 2012 were included in this study. Of these, 121 were positive for mycobacterial culture previously detected in our laboratory, comprising 100 MTB-positive specimens and 21 NTM-positive specimens. Nineteen specimens were MTB/NTM-negative. All cultures were performed using egg-based solid media (3% Ogawa medium, Asan Pharmaceutical Co., Seoul, Korea).
All sputum specimens were liquefied and decontaminated with N-acetyl-L-cysteine-2% NaOH and concentrated by centrifugation at 3,000g for 15 min. The sediment was resuspended in 1.5 mL of PBS.
All sputum specimens were subjected to rhodamine-auramine fluorochrome acid fast bacilli (AFB) staining, graded by using the United States Centers for Disease Control and Prevention (CDC)-recommended scale [9], and cultured in BacT/ALERT MP liquid medium and on 3% Ogawa solid medium. A positive culture isolate was rechecked by Ziehl-Neelsen staining, and identified by an immunochromatographic assay (BIOLINE SD TB Ag MPT64 Rapid, Standard Diagnostics, Yongin, Korea) and a real-time PCR assay (AdvanSure TB/NTM real-time PCR kit, LG Life Sciences, Seoul, Korea).
Twenty microliters of bacterial suspension were added to microscope slides. Each smear was air dried and heated at 80℃ for 2 hr. Smears were immersed in 80% ethanol for 30 min and subsequently air-dried and covered with approximately 20 µL of hybridization solution containing 20 mM Na2HPO4, pH 7.4, 20 mM Tris-HCl, pH 7.4, 60% formamide, 2×SSC solution, 0.1 µg/mL salmon sperm DNA (Biosesang, Seongnam, Korea), and 100 nM fluorescein-labeled PNA probe for MTB and NTM. The two probes were hybridized simultaneously to a single smear of the clinical sample. Slides were covered with coverslips and then placed in a heated moisture chamber in the dark at 55℃ for 1.5 hr. Following hybridization, the coverslips were removed, and the slides were submerged in pre-warmed 0.1% Triton X-100, 15 mM NaCl, and 5 mM Tris/HCl, pH 10 (Biosesang) in a moist chamber at 55℃ and washed for 30 min. The slides were then briefly immersed in distilled water. The sputum smears were finally mounted with one drop of Vysis DAPI IV mounting solution (Abbott Laboratories, Abbott Park, IL, USA) and covered with coverslips. Microscopic examination was undertaken by using a fluorescence microscope (Zeiss, Obercochen, Germany) equipped with a standard filter set. The results of PNA FISH were graded according to the United States CDC-recommended scale [9], same as for the routine fluorochrome AFB stain.
This research was reviewed and approved by the full committee review of the Institutional Review Board at Pusan National University Yangsan Hospital, Yangsan, Korea (no. 04-2013-023).
We evaluated the diagnostic sensitivity and specificity of PNA FISH by using the culture as the standard method. Agreement between tests was quantified using kappa statistics. The 95% confidence intervals (CI) of the kappa values were obtained by multiplying the Kappa variance by 1.96.
The MTB-specific PNA probe hybridized to rRNA of the tested MTB reference strain and did not cross-hybridize to rRNA of any of the NTM species tested. The NTM-specific PNA probe hybridized to rRNA of all the tested NTM species but did not hybridize to rRNA of the MTB reference strain. In a mixture of the MTB ATCC 13950 and M. kansasii ATCC 12479 strains, simultaneous identification of each species was possible (Fig. 1). Neither probe bound to the 10 frequently isolated respiratory bacteria, i.e., no cross-hybridization to any of the species was detected (Table 1).
The performance of the PNA FISH method was evaluated against culture as the gold standard. The MTB-specific PNA probe showed diagnostic sensitivity of 82% and diagnostic specificity of 100% (kappa=0.72, 95% CI: 0.607-0.837). The NTM-specific PNA probe showed diagnostic sensitivity of 71.4% and diagnostic specificity of 100% (kappa=0.81, 95% CI: 0.663-0.956) (Table 2). The percentages of false positives and false negatives were 0% and 18% for the MTB-specific probe, and 0% and 28.5% for the NTM-specific probe. The positive predictive and negative predictive values of a valid test result obtained with MTB PNA FISH were 100% and 44.2%, respectively.
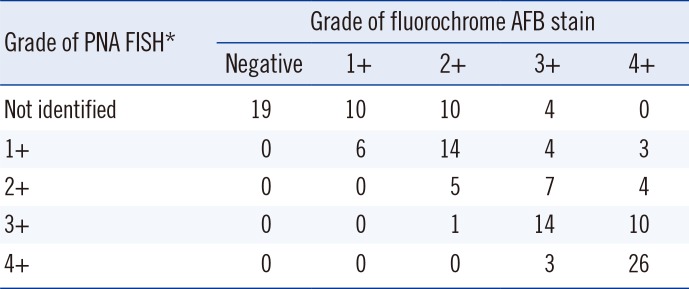
In total, 82.9% of the clinical specimens were correctly identified, with diagnostic sensitivity of 80.2% and diagnostic specificity of 100% (kappa=0.52, 95% CI 0.370-0.676). None of the samples was incorrectly identified as MTB or NTM, whereas 24 were negative with both probes and thus gave results that were inconclusive. Negative hybridization results were obtained for 18 MTB-positive specimens and six NTM-positive specimens identified by AFB stain. Their CDC scale fluorochrome AFB stain grades comprised nine specimens of 1+, nine specimens of 2+, and six specimens of 3+. None of the specimens was grade 4+ (Table 3). There were no false-positive or double-positive reactions.
Both MTB and NTM were observed as rod-shaped bacilli (Fig. 1). All cases were morphologically distinguishable from background artifacts.
The comparison of smear results obtained by the fluorochrome AFB stain and PNA FISH method is shown in Table 3. The correlation between the two methods was decreased in samples with lower bacillary counts. The difference between the techniques was particularly pronounced among grades lower than 2+.
Our PNA-probe-based FISH assay is a rapid and simple method for the detection of mycobacteria via smear microscopy. The use of PNA FISH probes targeting rRNA enables direct detection of single bacilli without any additional procedure [456]. The combined use of the MTB-specific and the NTM-specific probes within a single test gives a result regarding not only the MTB infection status but also the NTM infection status.
We expected the combined use of the MTB and NTM sequence-specific PNA FISH probes to ensure a very high positive predictive value and high sensitivity. However, the PNA FISH test results showed similar or lower sensitivity compared with conventional microscopic examination using direct clinical specimens. The sensitivity of conventional microscopy ranged from 32% to 94%, and the sensitivity of fluorescence microscopy ranged from 52% to 97% [10].
Such results for sensitivity may be obtained in cases of a few NTM species, which are not suitable for identification with the NTM-specific PNA probe used in this study because the NTM-specific probe does not cover all NTM species. Stenders et al. [6] reported false-negative results for a few NTM species; e.g., M. xenopi, M. fortuitum, and M. flavescens, which are not identified with the NTM specific PNA probe. The proportion of false-negative results for NTM will depend on the region of the world from which the test samples are derived. In Korea, the most frequently isolated organisms from clinical respiratory specimens are M. abscessus (29%), the M. fortuitum complex (17%), M. intracellulare (17%), and M. avium (15%) [11]; M. fortuitum complex is commonly found in Korea, however, it is not detectable with the probe using in the present study. Therefore the false-negative result of M. fortuitum complex could be a diagnostic problem in Korea.
A possible explanation for these false-negative results could be the loss of mycobacteria from the slide during the hybridization procedure. For optimal hybridization results, a large number of mycobacteria fixed on the smears are required to counteract the loss of bacteria during the hybridization procedure. Similar results were reported for a commercial PNA assay using material taken directly from broth cultures [4].
Low bacterial counts could be relevant here. Stender et al. [6] report a small proportion of negative results with both the MTB-specific and the NTM-specific PNA probes in cases for which smears contained only a small amount of material. Microscopic examination is insensitive, requiring 5,000-10,000 bacilli per milliliter of sputum for detection, and various studies indicate that only 50-80% of patients with untreated pulmonary TB have positive smears [9].
In previous studies, PNA-probe-based FISH assays were better able to differentiate between MTB and NTM in cultured mycobacterial isolates than in clinical samples [456]. In one study, the results obtained with 30 smears of cultured isolates were compared with AccuProbe mycobacterium identification kits (Hologic, Bedford, MA, USA), which revealed 84% diagnostic sensitivity for the MTB-specific PNA probe and 91% for the NTM-specific PNA probe. The specificity of both probes was 100%. Another study using 53 smears of Löwenstein-Jensen solid-medium cultures and 77 smears of Mycobacteria Growth Indicator Tube (MGIT, BD Biosciences, Franklin, NJ, USA) liquid cultures showed 98-99% diagnostic sensitivity, whereas the diagnostic sensitivity of the NTM probe was 57-100%. The specificity of each probe in both studies was 100%. The fact that a culture isolate contains a larger number of mycobacteria than a direct clinical specimen may explain this difference in sensitivity.
In the present study, we designed the PNA-FISH method not only for detection of mycobacteria but also for identification of MTB and NTM simultaneously in direct clinical specimens. O'Keefe et al. [5] published a study, in which multicolor multiplex PNA-FISH was used in mixed-organism populations for identification of various bacteria. In their work, mixtures of four organisms were simultaneously hybridized with four PNA probes, labeled with different fluorescent dyes. With this in mind, we designed the present study to use different fluorescence-labeled PNA probes for simultaneous identification of MTB and NTM on a single slide, for rapid and effective identification and differentiation of MTB and NTM. We observed that each organism simultaneously hybridized with two PNA probes labeled with different fluorescent dyes, and it was easy to differentiate them.
Detection and differentiation of mycobacteria require laborious and time-consuming biochemical techniques. Therefore, more rapid molecular techniques, based on nucleic-acid amplification and hybridization of the amplified product, were developed. However, these techniques require specific equipment and highly trained technicians. To overcome these problems, simple and rapid immunochromatographic assays were developed. Probe-detection methods that target rRNA are rapid and simple, giving results in 1-2 hr with an estimated accuracy above 90%, although they cover a limited range of species. The use of rapid identification tests in combination with rapid culture has greatly reduced overall turnaround times in clinical laboratories.
The TB PNA-FISH test provides a combination of the high specificity offered by molecular techniques and easy adaptability to most clinical laboratories. The molecular technique, hydrophobic PNA, and specifically designed probes, give high specificity. Furthermore, the TB PNA-FISH test reveals the bacterial morphology as an additional means of identification, which can contribute to higher specificity. The test uses standard equipment available in most clinical laboratories and does not require any specialized equipment. It is simple to perform, so highly trained technicians are not required. The test requires 5-6 hr to complete, and the results can be reported within a day. The low cost per test, including reagents and consumables, is also an advantage of this method. Compared with molecular techniques, this method is simpler, faster, and cheaper, while providing a similar specificity.
The relatively low sensitivity could be a weak point of this method. In the clinical laboratory, the TB PNA-FISH test needs optimization for higher sensitivity. When using the PNA-FISH method for detection and identification in a direct specimen with low bacterial counts, unidentified specimens were subjected to further workup. The PNA FISH method is applicable to specimens with high bacterial counts, such as cultures.
In conclusion, the PNA-FISH assay is a useful method for rapid and accurate identification of mycobacteria and is easily adaptable to current microscopic techniques that are routinely used in the majority of clinical microbiology laboratories. It is a new stain-based method that can differentiate MTB and NTM. Using our procedure, the results obtained with the MTB-specific and NTM-specific PNA probes showed good diagnostic specificity and agreement with the results obtained by other diagnostic methods, especially in specimens with high bacterial counts.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012R1A1A2004593).
References
1. World Health Organization. Global tuberculosis report 2014. Geneva: World Health Organization;2014.
2. Shinnick TM, Iademarco MF, Ridderhof JC. National plan for reliable tuberculosis laboratory services using a systems approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR Recomm Rep. 2005; 54:1–12. PMID: 15829862.
3. Fitzgerald DW, Sterling TR, Hass DW. Mycobacterium tuberculosis. In : Mandell GL, Bennett JE, Dolan R, editors. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. 7th ed. Philadelphia, PA: Elsevier;2010.
4. Padilla E, Manterola JM, Rasmussen OF, Lonca J, Domínguez J, Matas L, et al. Evaluation of a fluorescence hybridisation assay using peptide nucleic acid probes for identification and differentiation of tuberculous and non-tuberculous mycobacteria in liquid cultures. Eur J Clin Microbiol Infect Dis. 2000; 19:140–145. PMID: 10746504.

5. Perry-O'Keefe H, Rigby S, Oliveira K, Sørensen D, Stender H, Coull J, et al. Identification of indicator microorganisms using a standardized PNA FISH method. J Microbiol Methods. 2001; 47:281–292. PMID: 11714518.
6. Stender H, Lund K, Petersen KH, Rasmussen OF, Hongmanee P, Miörner H, et al. Fluorescence In situ hybridization assay using peptide nucleic acid probes for differentiation between tuberculous and nontuberculous mycobacterium species in smears of mycobacterium cultures. J Clin Microbiol. 1999; 37:2760–2765. PMID: 10449448.

7. Porcheddu A, Giacomelli G. Peptide nucleic acids (PNAs), a chemical overview. Curr Med Chem. 2005; 12:2561–2599. PMID: 16248816.

8. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215:403–410. PMID: 2231712.

9. Diagnostic Standards and Classification of Tuberculosis in Adults and Children. This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am J Respir Crit Care Med. 2000; 161:1376–1395. PMID: 10764337.
10. Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, Cunningham J, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006; 6:570–581. PMID: 16931408.

11. Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006; 129:341–348. PMID: 16478850.

Fig. 1
Peptide nucleic acid (PNA) FISH analysis. (A) Positive PNA FISH for Mycobacterium tuberculosis (MTB). (B) Positive PNA FISH for non-tuberculous mycobacteria (NTM). (C) PNA FISH for a mixture of the MTB ATCC 13950 and M. kansasii ATCC 12479 strains. The MTB PNA probe was labeled with 6-carboxyfluorescein fluorescent dye (green, filled arrows), and the NTM PNA probe was labeled with cyanine 3 fluorescent dye (orange, non-filled arrows).

Table 1
Results of PNA FISH using MTB- and NTM-specific probes for frequently isolated mycobacteria and other bacteria in clinical laboratory
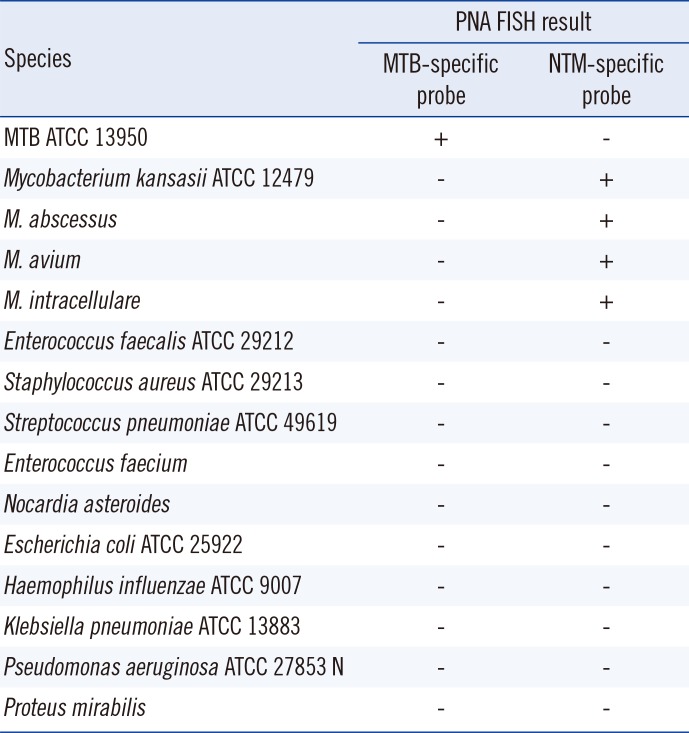
Table 2
Comparison of the PNA FISH assay with culture

| PNA FISH | Culture | ||
|---|---|---|---|
| MTB-positive | NTM-positive | MTB/NTM-negative | |
| MTB-positive | 82 | 0 | 0 |
| NTM-positive | 0 | 15 | 0 |
| MTB/NTM-not identified | 18 | 6 | 19 |
| Total | 100 | 21 | 19 |
Table 3
Comparison of results obtained by PNA FISH assay and fluorochrome AFB stain
| Grade of PNA FISH* | Grade of fluorochrome AFB stain | ||||
|---|---|---|---|---|---|
| Negative | 1+ | 2+ | 3+ | 4+ | |
| Not identified | 19 | 10 | 10 | 4 | 0 |
| 1+ | 0 | 6 | 14 | 4 | 3 |
| 2+ | 0 | 0 | 5 | 7 | 4 |
| 3+ | 0 | 0 | 1 | 14 | 10 |
| 4+ | 0 | 0 | 0 | 3 | 26 |
*Graded by using the United States Centers for Disease Control and Prevention-recommended scale [9].
Abbreviations: PNA, peptide nucleic acid; AFB, acid-fast bacilli.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download