 | Fig. 1Bone marrow karyotype showing derivative chromosomes 10 and 12 with an apparently normal chromosome 11. Karyotype: 46,XY,der(10)t(10;21)(p12;q21),der(12)del(12)(p11.2)add(12)(q24.2)[17]/46,XY[3]. |
Dear Editor
The mixed lineage leukemia gene (MLL) on chromosome 11q23 is a frequent target of chromosomal translocations and rearrangements in childhood leukemia and adult therapy-related leukemia [1]. By using a commercially available dual-color FISH break-apart probe, almost all MLL rearrangements can be detected, including variant and cryptic forms [2, 3, 4]. We report a unique case of cryptic MLL rearrangement, in which the patient exhibited a karyotype that did not include t(10;11)(p12;q23) and FISH results did not reveal MLL break-apart signals; however, molecular analysis by reverse transcription (RT)-PCR revealed a 388-bp amplicon corresponding to the MLL/MLLT10 fusion transcript.
A 17-yr-old male was admitted to our hospital for evaluation and management of multiple palpable neck lymphadenopathies. Blasts accounted for approximately 94% of cells on peripheral blood smear. On bone marrow (BM) aspiration smear, 90.5% of nucleated cells were blasts.
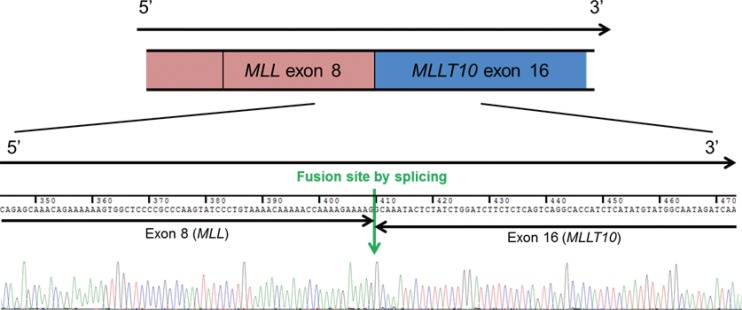
Chromosomes were analyzed via GTG banding. Complex structural abnormalities were observed, and the karyotype was 46,XY,der(10)t(10;21)(p12;q21),der(12)del(12)(p11.2)add(12)(q24.2)[17]/46,XY[3] (Fig. 1). FISH analysis was performed according to the manufacturer's instructions, including MLL (LSI Dual-Color, Break Apart Rearrangement Probe, Vysis, Downers Grove, IL, USA). No break-apart signals were observed for the MLL probe (Fig. 2). Multiplex RT-PCR was performed with HemaVision (DNA Technology, Aarhus, Denmark), according to the manufacturer's instructions. Molecular analysis revealed an amplicon corresponding to the MLL/MLLT10 fusion transcript. RT-PCR analysis was performed with a combination of 5' MLL and 3' MLLT10 primers: MLL Forward 5'-GGAAGTCAAG CAAGCAGGTC-3' and MLLT10 Reverse 5'-GCTGCCATTGATGAATTTG-3'. PCR products were subjected to direct sequencing by using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) on ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). RT-PCR revealed a 386-bp product ranging from the exon 8 region of the MLL gene to the exon 16 region of the MLLT10 gene, linked via two orphan nucleotides (Fig. 3).
A diagnosis of "variant MLL translocation in acute leukemia" was made. The patient was treated with induction chemotherapy, including cytarabine and idarubicin. Complete hematologic remission was achieved 24 days later.
Conventional cytogenetic analysis can miss MLL rearrangements, possibly owing to poor metaphase division of leukemic cells and complex or cryptic chromosomal abnormalities. FISH and RT-PCR are advantageous because they are fast and target specific genes [5]. However, most commonly employed multiplex RT-PCR kits detect a limited number of rearrangements, since only the most common regions are included [5, 6, 7]. MLL rearrangement cases outside of the breakpoint cluster region can be detected by FISH despite being RT-PCR negative [8]. Currently, long-distance inverse PCR (LDI-PCR) is the most powerful, verified method for detecting known and unknown partner genes in MLL rearrangements [8, 9]. In our case, a genetic aberration, an MLL (exon 8)-MLLT10 (exon 16) rearrangement, was confirmed by sequencing. However, we were not able to perform additional testing such as chromosome microarrays or re-testing with different FISH probes (i.e., from a different manufacturer) or LDI-PCR because of insufficient specimens.
The MLLT10/AF10 gene was found to be involved in "chromosomal fragment insertions," a complex mechanism that occurs when either a fragment of chromosome 11 (including portions of the MLL gene) is inserted into another chromosome, or a fragment of another chromosome (including portions of a translocation partner gene) is inserted into the breakpoint cluster region of the MLL gene, and is most frequently involved in the latter mechanism [9]. The MLLT10/AF10 gene was also found to be one of the 20 genes that are involved in complex rearrangements, with three- or four-way translocations resulting in more than two fusion alleles [9]. While the 3'-portions of the translocation partner genes were regularly fused to the 5'-portion of the MLL gene, the genes involved in complex rearrangements, including the MLLT10/AF10 gene, were fused to the 3' portion of the MLL gene [9]. There is a high likelihood that our case also involved a complex rearrangement, considering that the 3' portion of the MLL gene was fused to the 5' portion of the MLLT10 gene.
To the best of our knowledge, this is the first case report of a cryptic MLL rearrangement detected by RT-PCR only, despite testing negative for FISH. However, this case may not represent a truly cryptic MLL rearrangement, considering that the chromosome results revealed a complex karyotype. While FISH assays are recognized for their usefulness in detecting cryptic MLL rearrangements, this case report emphasizes that RT-PCR can be useful for detecting FISH-negative cryptic MLL rearrangements. It would be useful to document other similar cases so that a strategy for diagnosis can be developed.
Notes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
Go to : 
References
1. de Boer J, Walf-Vorderwülbecke V, Williams O. In focus: MLL-rearranged leukemia. Leukemia. 2013; 27:1224–1228. PMID: 23515098.

2. Morerio C, Rapella A, Rosanda C, Lanino E, Lo Nigro L, Di Cataldo A, et al. MLL-MLLT10 fusion in acute monoblastic leukemia: variant complex rearrangements and 11q proximal breakpoint heterogeneity. Cancer Genet Cytogenet. 2004; 152:108–112. PMID: 15262427.

3. Stasevich I, Utskevich R, Kustanovich A, Litvinko N, Savitskaya T, Chernyavskaya S, et al. Translocation (10;11)(p12;q23) in childhood acute myeloid leukemia: incidence and complex mechanism. Cancer Genet Cytogenet. 2006; 169:114–120. PMID: 16938568.

4. Van Limbergen H, Poppe B, Janssens A, De Bock R, De Paepe A, Noens L, et al. Molecular cytogenetic analysis of 10;11 rearrangements in acute myeloid leukemia. Leukemia. 2002; 16:344–351. PMID: 11896537.

5. Yang JJ, Marschalek R, Meyer C, Park TS. Diagnostic usefulness of genomic breakpoint analysis of various gene rearrangements in acute leukemias: a perspective of long distance- or long distance inverse-PCR-based approaches. Ann Lab Med. 2012; 32:316–318. PMID: 22779077.

6. Choi HJ, Kim HR, Shin MG, Kook H, Kim HJ, Shin JH, et al. Spectra of chromosomal aberrations in 325 leukemia patients and implications for the development of new molecular detection systems. J Korean Med Sci. 2011; 26:886–892. PMID: 21738341.

7. Kim MJ, Choi JR, Suh JT, Lee HJ, Lee WI, Park TS. Diagnostic standardization of leukemia fusion gene detection system using multiplex reverse transcriptase-polymerase chain reaction in Korea. J Korean Med Sci. 2011; 26:1399–1400. author reply 1401. PMID: 22022200.
8. Watanabe N, Kobayashi H, Ichiji O, Yoshida MA, Kikuta A, Komada Y, et al. Cryptic insertion and translocation or nondividing leukemic cells disclosed by FISH analysis in infant acute leukemia with discrepant molecular and cytogenetic findings. Leukemia. 2003; 17:876–882. PMID: 12750700.

9. Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013; 27:2165–2176. PMID: 23628958.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download