Abstract
Background
Data on allele frequencies (AFs) and haplotype frequencies (HFs) of HLA-C and -DQB1 are limited in Koreans. We investigated AFs and HFs of HLA-A, -B, -C, -DRB1, and -DQB1 in Koreans by high-resolution sequence-based typing (SBT).
Methods
Hematopoietic stem cells were obtained from 613 healthy, unrelated donors to analyze HLA-A, -B, -C, -DRB1, and -DQB1 genotypes by using AlleleSEQR HLA-A, -B, -C, -DRB1, and -DQB1 SBT kits (Abbott Molecular, USA), respectively. Alleles belonging to HLA-C*07:01/07:06 group were further discriminated by using PCR-sequence specific primer analysis. AFs and HFs were calculated by direct counting and maximum likelihood method, respectively.
Results
In all, 24 HLA-A, 46 HLA-B, 24 HLA-C, 29 HLA-DRB1, and 15 HLA-DQB1 alleles were identified. AFs and HFs of HLA-A, -B, and -DRB1 were similar to those reported previously. For the HLA-C locus, C*01:02 was the most common allele, followed by C*03:03, C*03:04, C*14:02, C*03:02, and C*07:02 (AF ≥7%). AFs of C*07:01 and C*07:06 were 0.16% and 3.18%, respectively. For the HLA-DQB1 locus, DQB1*03:01 was the most common allele, followed by DQB1*03:03, *03:02, *06:01, *05:01, *04:01, and *06:02 (AF ≥7%). AFs of DQB1*02:01 and DQB1*02:02 were 2.12% and 6.69%, respectively. HFs of A*33:03-C*07:06 and C*07:06-B*44:03 were 3.09% and 3.10%, respectively, while those of DRB1*07:01-DQB1*02:02 and DRB1*03:01-DQB1*02:01 were 6.61% and 2.04%, respectively.
Number of known alleles has rapidly increased because of the use of DNA-based HLA typing methods. Allele (AF) and haplotype frequencies (HF) of HLA are different among different ethnic groups and regions. In Koreans, AF and HF of HLA are reported by using generic DNA typing [1, 2, 3, 4] and high-resolution genotyping [5, 6, 7]. However, genotyping data for both of HLA-C and -DQB1 alleles using sequence-based typing (SBT) have not been obtained in previous studies, and some alleles such as C*07:01/*07:06 and DQB1*02:01/02:02 have not been discriminated [2, 3, 4, 5]. Recent studies have shown that HLA-C and -DQB1 matches are important for hematopoietic stem cell and organ transplantations [8, 9, 10, 11, 12]. Understanding the exact frequencies of HLA and other common, well-defined alleles in different ethnic groups is important to resolve ambiguous results of HLA typing. Higher AF of C*07:06 in the Chinese than in Caucasians was the cause of PCR dropout of widely used AlleleSEQR HLA-C Plus kit (Abbott Molecular, Des Plaines, IL, USA) because the kit was mainly developed for HLA typing for Caucasians [13]. The present study analyzed the AF and HF of HLA-A, -B, -C, -DRB1, and -DQB1 in Koreans by using high-resolution SBT and compared them with other ethnic groups.
Hematopoietic stem cells were obtained from 613 healthy Korean donors (559 healthy unrelated adults and 54 umbilical cord blood units) for high-resolution HLA typing at Seoul National University Hospital from January 2006 to July 2014. This study was approved by the Institutional Review Board (IRB) of Seoul National University Hospital (IRB No. 1408-028-601).
Genomic DNA was extracted from white blood cells obtained from the peripheral blood of the donors and umbilical cord blood units by using QuickGene-Mini80 DNA isolation system (Fujifilm, Tokyo, Japan).
SBT of HLA-A, -B, -C, -DRB1, and -DQB1 was performed by using AlleleSEQR HLA-A, -B, -C, -DRB1, and -DQB1 SBT kits (Abbott Molecular), respectively. For HLA class I alleles (HLA-A, -B, and -C), PCR mixture was prepared using 16 µL of master mix, 80-160 ng of genomic DNA, and 0.3 µL of AmpliTaq Gold polymerase (Abbott Molecular) at a final volume of 10 µL. For HLA class II alleles (HLA-DRB1 and -DQB1), PCR mixture was prepared by using 8 µL of master mix, 40-80 ng of genomic DNA, and 0.1 µL of AmpliTaq Gold polymerase at a final volume of 10 µL. Amplification conditions were as follows: initial denaturation at 95℃ for 10 min, followed by 36 cycles of denaturation at 96℃ for 20 sec, annealing at 60℃ for 30 sec, and elongation at 72℃ for 3 min.
Next, 16 µL of the PCR products were mixed with 3 µL of ExoSAP-IT and were purified at 37℃ for 15-30 min and 80℃ for 15 min. Exons 2, 3, and 4 were sequenced for HLA class I alleles (HLA-A, -B, and -C); exon 2 and codon 86 were sequenced for HLA-DRB1; and exons 2 and 3 were sequenced for HLA-DQB1. Next, 8 µL of sequencing mix was added to 2 µL of the purified PCR products, and sequencing was performed by using 25 thermal cycles of denaturation at 96℃ for 20 sec, annealing at 50℃ for 30 sec, and elongation at 60℃ for 2 min. Next, 2 µL of sodium acetate/EDTA buffer and 25 µL of absolute ethanol (EtOH) were added to the mixture containing sequenced products. The mixture was then vigorously vortexed and centrifuged at 2,000g for 30 min, and the supernatant was removed. Next, 50 µL of 80% EtOH was added, and the mixture was centrifuged at 2,000g for 5 min; this process was repeated twice. Finally, 15 µL of highly deionized formamide (Applied Biosystems, Foster City, CA, USA) and 15 µL of 0.3 mM EDTA were added, and the mixture was loaded onto 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
The obtained electropherograms were processed by using Assign SBT (Conexio Genomics, Fremantle Western Australia, Australia). To resolve ambiguity in C*07:01/07:06/07:18 group, exon 5 sequences of HLA-C were further analyzed, as described previously [14].
AFs and HFs of the HLA alleles were determined by using direct counting and maximum likelihood method, respectively. For 2-locus haplotypes, relative linkage disequilibrium (RLD) values between two alleles at different loci were analyzed and chi-square values were calculated for statistical significance. HFs of HLA A-C-B-DRB1-DQB1 haplotypes and AFs of HLA-C and -DQB1 in Koreans were compared with those in other populations by using chi-square test or Fisher's exact test, as appropriate. Bonferroni correction was applied by multiplying P values with the number of comparisons made (24 for HLA A-C-B-DRB1-DQB1 haplotypes, 24 for HLA-C, and 14 for HLA-DQB1) to calculate corrected P values (Pc). Pc value of <0.05 was considered significantly different. All analyses were performed by using R version 3.1.2 software with gap package (http://cran.us.r-project.org/) [15].
AFs of HLA classes I and II are listed in Table 1. For the HLA-A locus, A2 group comprised more than 50% of all the HLA-A alleles. In all, 24 distinct alleles were identified for the HLA-A locus. Of these, A*24:02 was the most common allele, followed by A*02:01, A*33:03, A*02:06, and A*11:01 (AF ≥5%). The HLA-B locus showed the greatest diversity, with 46 alleles. Of these, B*51:01, B*15:01, and B*44:03 were the most common alleles, followed by B*58:01, B*35:01, B*54:01, and B*46:01 (AF ≥5%). In all, 24 alleles were identified for the HLA-C locus. Of these, C*01:02 was the most common allele, followed by C*03:03, C*03:04, C*14:02, C*03:02, C*07:02, C*08:01, C*06:02, and C*14:03 (AF ≥5%). AFs of C*07:01 and C*07:06 were 0.16% and 3.18%, respectively. In all, 29 alleles were identified for the HLA-DRB1 locus. Of these, DRB1*09:01 was the most common allele, followed by DRB1*13:02, DRB1*04:05, DRB1*15:01, DRB1*07:01, DRB1*08:03, and DRB1*01:01 (AF ≥5%). In all, 15 alleles were identified for the HLA-DQB1 locus. The sum of AFs of DQB1*03:01 and DQB1*03:03 were 25.2%. AFs of three DQ6 alleles (DQB1*06:01, DQB1*06:02, and DQB1*06:04), DQB1*02:02, DQB1*03:02, DQB1*04:01, and DQB1*05:01 were ≥5%. AFs of DQB1*02:01 and DQB1*02:02 were 2.12% and 6.69%, respectively.
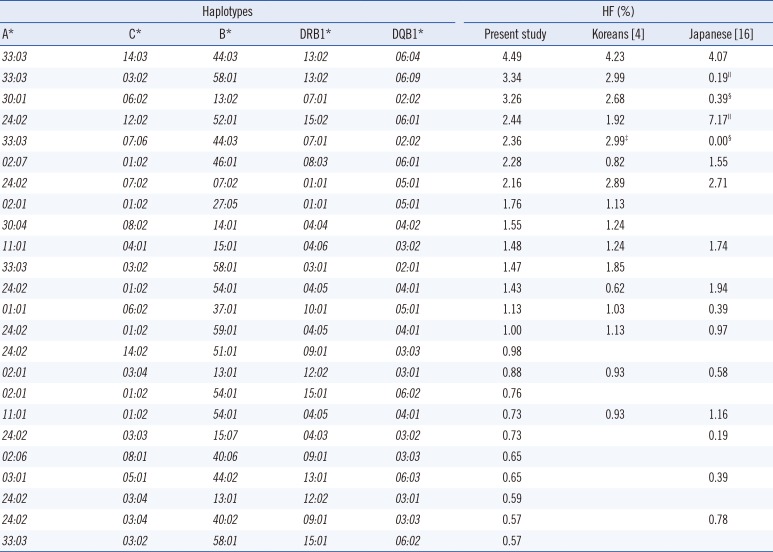
The 2-locus haplotypes exhibiting HFs higher than 0.9% are listed in Table 2. HFs of 31 A-C, 27 B-DRB1, 29 C-B, and 24 DRB1-DQB1 haplotypes were >0.9%. In each set, A*33:03-C*03:02, B*44:03-DRB1*13:02, C*14:02-B*51:01, and DRB1*09:01-DQB1*03:03 were the most common alleles. Three haplotypes (C*08:02-B*14:01, C*05:01-B*44:02, and DRB1*13:01-DQB1*06:03) showed the strongest associations with RLD values of >0.99. HFs (RLD, χ2) of A*33:03-C*07:06 and C*07:06-B*44:03 were 3.09% (0.40, 83.6) and 3.10% (0.59, 301.3), respectively, while those of DRB1*07:01-DQB1*02:02 and DRB1*03:01-DQB1*02:01 were 6.61% (0.97, 377.8) and 2.04% (0.96, 320.0), respectively. Table 3 lists 5-locus HLA A-C-B-DRB1-DQB1 haplotypes having HFs of >0.5%. The most common 5-locus haplotype was A*33:03-C*14:03-B*44:03-DRB1*13:02-DQB1*06:04, followed by A*33:03-C*03:02-B*58:01-DRB1*13:02-DQB1*06:09 (Table 3). HF of A*33:03-C*07:06-B*44:03-DRB1*07:01-DQB1*02:02 was 2.36%. HF of A*33:03-C*03:02-B*58:01-DRB1*13:02-DQB1*06:09 was higher while that of A*24:02-C*12:02-B*52:01-DRB1*15:02-DQB1*06:01 was lower than that in the Japanese (Pc<0.01) (Table 3).
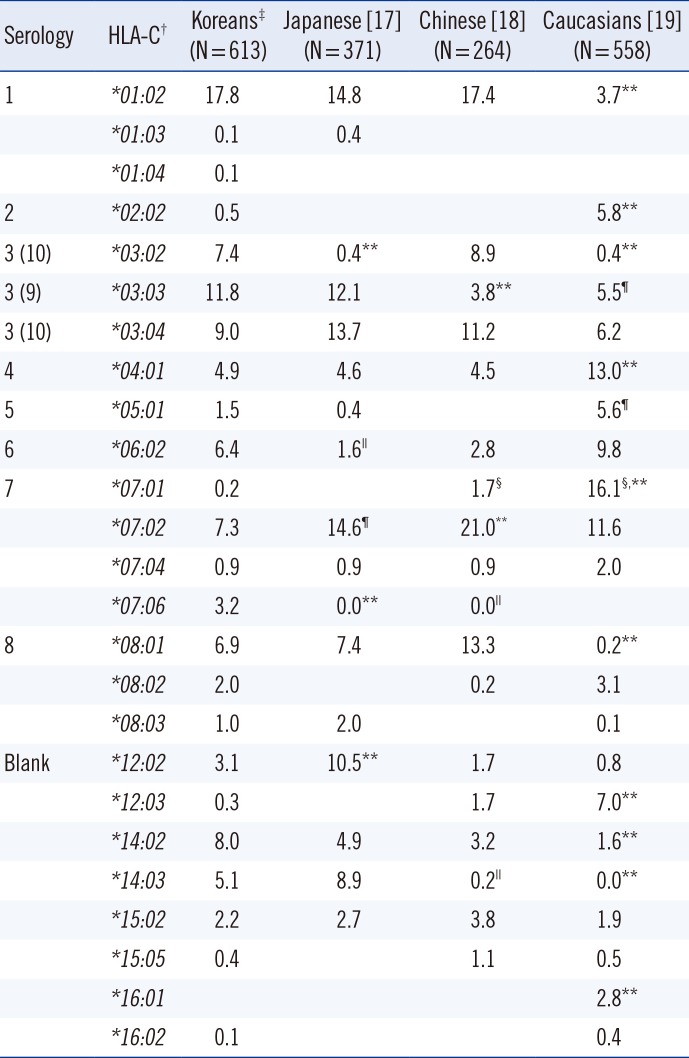
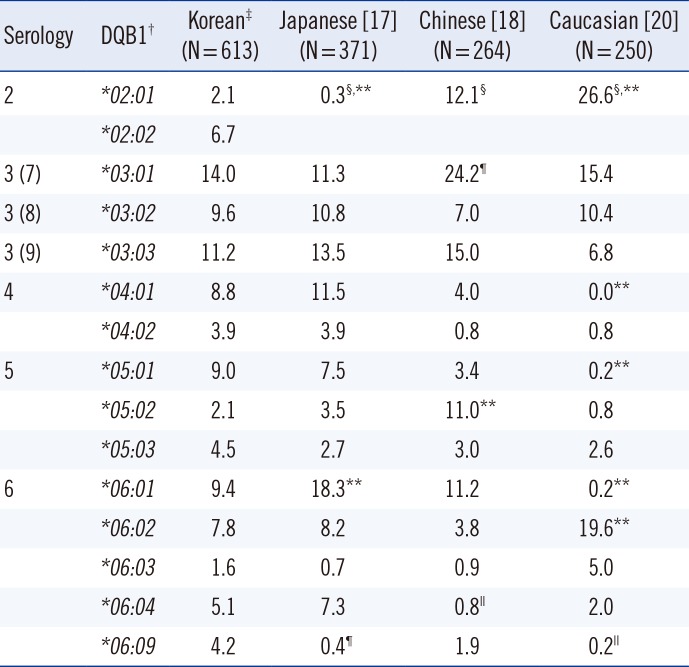
For the HLA-C locus, AFs of C*03:02 and C*07:01/06 were higher and that of C*12:02 was lower than those in the Japanese (Pc<0.001; Table 4). AF of C*03:03 was higher and that of C*07:02 was lower than that in the Chinese (Pc<0.001). AFs of C*01:02, C*03:02, C*08:01, C*14:02, and C*14:03 were higher and those of C*02:02, C*04:01, C*07:01/06, C*12:03, and C*16:01 were lower than those in Caucasians (Pc<0.001; Table 4). For the HLA-DQB1 locus, AF of DQB1*02:01/02 was higher and that of DQB1*06:01 was lower than that in the Japanese (Pc<0.001; Table 5). AF of DQB1*05:02 was lower than that in the Chinese (Pc<0.001). AFs of DQB1*04:01, *05:01, and *06:01 were higher and those of DQB1*02:01/02 and DQB1*06:02 were lower than those in Caucasians (Pc<0.001; Table 5).
In this study, we investigated the distribution of AFs and HFs of five HLA loci (HLA-A, -B, -C, -DRB1, and -DQB1) in 613 healthy Korean hematopoietic stem cell donors by using high-resolution DNA typing. AFs of HLA-A, -B, -DRB1, and -DQB1 were similar to those reported in previous studies [1, 2, 3, 4, 5, 6, 7]. For the HLA-C locus, AF of C*03:02 (7.42%) was lower but that of C*03:04 (8.97%) was higher than that determined by using PCR-sequence specific oligonucleotides (PCR-SSO) and sequencing in a previous study involving 485 healthy Koreans (10.82% for C*03:02 and 3.82% for C*03:04) [4]. However, AFs reported in our study were similar to those obtained by using PCR-SSO and sequence-specific conformational polymorphism in another study involving 474 healthy Koreans (6.3% for C*03:02 and 9.9% for C*03:04) [5]. This discrepancy should be clarified in further studies by performing high-resolution DNA typing in a larger study population.
Compared with the previous two studies [4, 5], we further discriminated HLA-C*07:01/*07:06 and determined AFs of C*07:01 (0.16%) and C*07:06 (3.18%) and HFs of A*33:03-C*07:06 (3.09%) and C*07:06-B*44:03 (3.10%), with significant χ2 values (83.6 and 301.3, respectively). Strong linkage disequilibrium of C*07:06 with A*33:03 or B*44:03 observed in the present study was in accordance with that reported in a previous study involving 118 healthy Koreans [14]. Therefore, most A*33:03-C*07:01/06 and C*07:01/06-B*44:03 haplotypes reported previously [4, 5] would be A*33:03-C*07:06 and C*07:06-B*44:03 [4, 5]. Further, we discriminated DQB1*02:01/*02:02 and determined AFs of DQB1*02:01 (2.12%) and DQB1*02:02 (6.69%) and HFs of DRB1*03:01-DQB1*02:01 (2.04%) and DRB1*07:01-DQB1*02:02 (6.61%), with significant χ2 values (320.0 and 377.8, respectively), which were similar to those reported in a previous study involving 414 parents of 207 Korean families [2]. For the 5-locus haplotypes, the results of the present study implied that most A*33:03-C*07:01/06-B*44:03-DRB1*07:01-DQB1*02:01/02 haplotype reported previously, with HF of 2.99% [4], would be A*33:03-C*07:06-B*44:03-DRB1*07:01-DQB1*02:02.
In the present study, all the DQB1 alleles had AFs of >1.5%, with no additional DQB1 allele, compared with those reported in previous studies [2, 4], indicating lower polymorphism of the HLA-DQB1 locus in Koreans. Thus, for organ transplantation, HLA-DQB1 alleles might be analyzed by using an intermediate resolution typing kit to determine compatible donors or to define donor-specific HLA antibody.
AFs of HLA-C alleles in different ethnic groups are listed in Table 4. From the perspective of killer cell immunoglobulin-like receptor (KIR) ligand status, the portion of HLA-C1 group (HLA-C alleles expressing Asn80, such as HLA-Cw1, -Cw3, -Cw7, -Cw8, -Cw12, -Cw13, -Cw14, and -Cw16) [21] was higher in Koreans than in Caucasians (84.2% vs. 61.1%). The portion of C2 group (HLA-C alleles expressing Lys80, such as HLA-Cw2, -Cw4, -Cw5, -Cw6, -Cw15, -Cw17, and -Cw18) was lower in Koreans than in Caucasians (15.8% vs. 36.6%). These differences could affect the frequency of KIR ligand mismatch for hematopoietic stem cell transplantation and possibly the clinical outcome. AFs of HLA-DQB1 alleles in different ethnic groups are listed in Table 5. AF of DQB1*06:01 was higher in Koreans (9.4%) than in Caucasians (0.2%), which might cause false reaction while using commercial kits developed in the United States [22].
In summary, we determined the AFs and HFs of HLA-A, -B, -C, -DRB1, and -DQB1 in Koreans by using high-resolution SBT. These data could be used to determine exact HLA types in Koreans for organ and hematopoietic stem cell transplantations.
References
1. Roh EY, Kim HS, Kim SM, Lim YM, Han BY, Park MH. HLA-A, -B, -DR allele frequencies and haplotypic associations in Koreans defined by generic-level DNA typing. Korean J Lab Med. 2003; 23:420–430.
2. Song EY, Park MH, Kang SJ, Park HJ, Kim BC, Tokunaga K, et al. HLA class II allele and haplotype frequencies in Koreans based on 107 families. Tissue Antigens. 2002; 59:475–486. PMID: 12445317.

3. Hwang SH, Oh HB, Yang JH, Kwon OJ, Shin ES. Distribution of HLA-A, B, C allele and haplotype frequencies in Koreans. Korean J Lab Med. 2004; 24:396–404.
4. Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens. 2005; 65:437–447. PMID: 15853898.

5. Chung HY, Yoon JA, Han BY, Song EY, Park MH. Allelic and haplotypic diversity of HLA-A, -B, -C, and -DRB1 genes in Koreans defined by high-resolution DNA typing. Korean J Lab Med. 2010; 30:685–696. PMID: 21157157.

6. Huh JY, Yi DY, Eo SH, Cho H, Park MH, Kang MS. HLA-A, -B and -DRB1 polymorphism in Koreans defined by sequence-based typing of 4128 cord blood units. Int J Immunogenet. 2013; 40:515–523. PMID: 23724919.

7. Yoon JH, Shin S, Park MH, Song EY, Roh EY. HLA-A, -B, -DRB1 allele frequencies and haplotypic association from DNA typing data of 7096 Korean cord blood units. Tissue Antigens. 2010; 75:170–173. PMID: 20196826.

8. Petersdorf EW, Gooley T, Malkki M, Horowitz M. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Clinical significance of donor-recipient HLA matching on survival after myeloablative hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007; 69(Suppl 1):25–30. PMID: 17445158.

9. Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007; 110:4576–4583. PMID: 17785583.

10. Tiercy JM. HLA-C incompatibilities in allogeneic unrelated hematopoietic stem cell transplantation. Front Immunol. 2014; 5:216. PMID: 24904572.

11. Aubert O, Bories MC, Suberbielle C, Snanoudj R, Anglicheau D, Rabant M, et al. , Risk of antibody-mediated rejection in kidney transplant recipients with anti-HLA-C donor-specific antibodies. Am J Transplant. 2014; 14:1439–1445. PMID: 24804568.
12. Willicombe M, Brookes P, Sergeant R, Santos-Nunez E, Steggar C, Galliford J, et al. De novo DQ donor-specific antibodies are associated with a significant risk of antibody-mediated rejection and transplant glomerulopathy. Transplantation. 2012; 94:172–177. PMID: 22735711.

13. Deng Z, Wang D, Xu Y, Gao S, Zhou H, Yu Q, et al. HLA-C polymorphisms and PCR dropout in exons 2 and 3 of the Cw*0706 allele in sequence-based typing for unrelated Chinese marrow donors. Hum Immunol. 2010; 71:577–581. PMID: 20226825.

14. Lee KW, Jung YA. Refined information on alleles belonging to the C*07:01/07:06/07:18 group in the Korean population. Human Immunol. 2011; 72:723–726. PMID: 21663780.

15. The Comprehensive R Archive Network. Updated on Oct 2014. http://cran.us.r-project.org/.
16. Nakajima F, Nakamura J, Yokota T. Analysis of HLA haplotypes in Japanese, using high resolution allele typing. MHC. 2001; 8:1–32.

17. Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 2000; 56:522–529. PMID: 11169242.

18. Trachtenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, Erlich H, et al. HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China. Tissue Antigens. 2007; 70:455–463. PMID: 17900288.

19. Mack SJ, Tu B, Lazaro A, Yang R, Lancaster AK, Cao K, et al. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens. 2009; 73:17–32. PMID: 19000140.

20. Dunne C, Crowley J, Hagan R, Rooney G, Lawlor E. HLA-A, B, Cw, DRB1, DQB1 and DPB1 alleles and haplotypes in the genetically homogenous Irish population. Int J Immunogenet. 2008; 35:295–302. PMID: 18976432.

21. Ruggeri L, Mancusi A, Burchielli E, Aversa F, Martelli MF, Velardi A. Natural killer cell alloreactivity in allogeneic hematopoietic transplantation. Curr Opin Oncol. 2007; 19:142–147. PMID: 17272987.

22. Roh EY, In JW, Shin S, Yoon JH, Park KU, Song EY. Performance of LIFECODES HLA-DQB1 typing kit using Luminex platform in Koreans. Ann Lab Med. 2015; 35:123–127. PMID: 25553292.

Table 1
AFs (%) of HLA-A, -B, -C, -DRB1, and -DQB1 in Koreans (N=613)
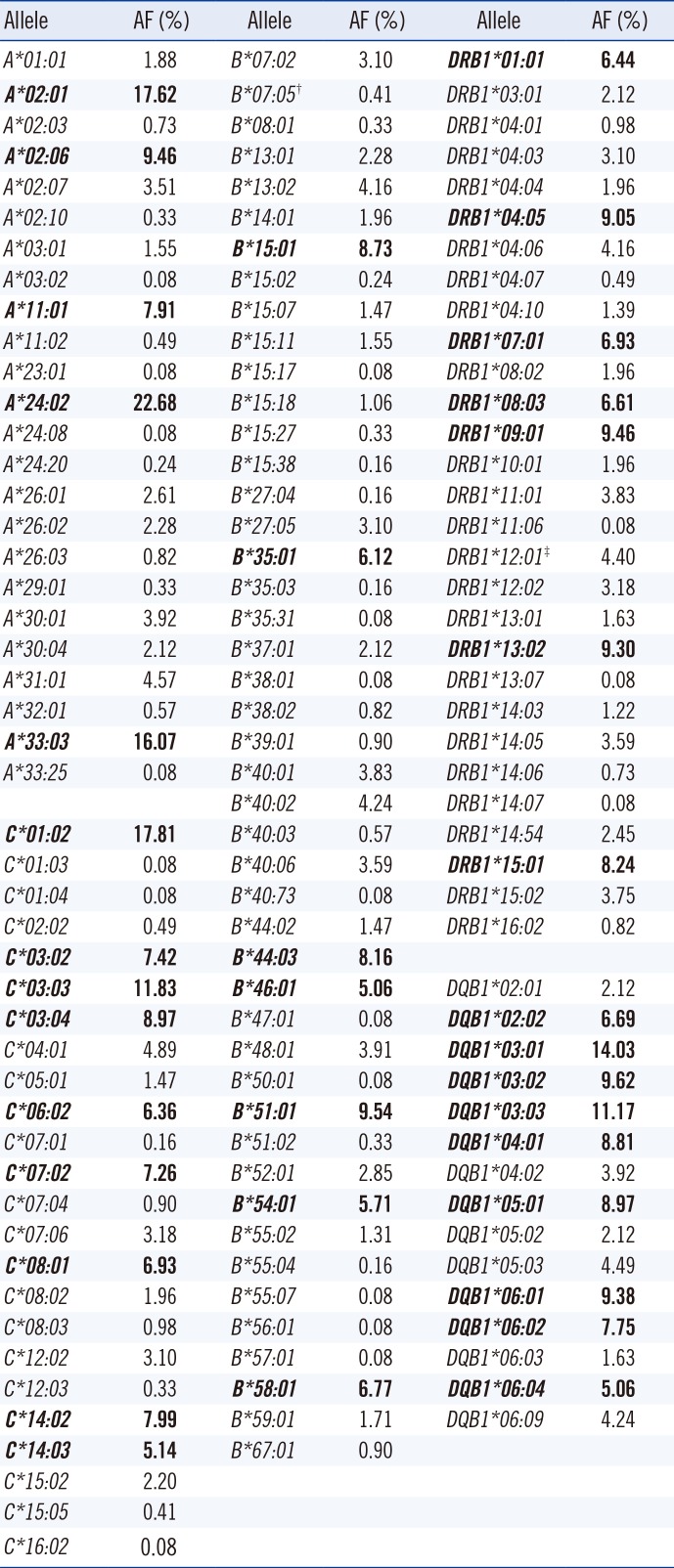
Table 2
HFs (%) of two-locus HLA haplotypes in Koreans†
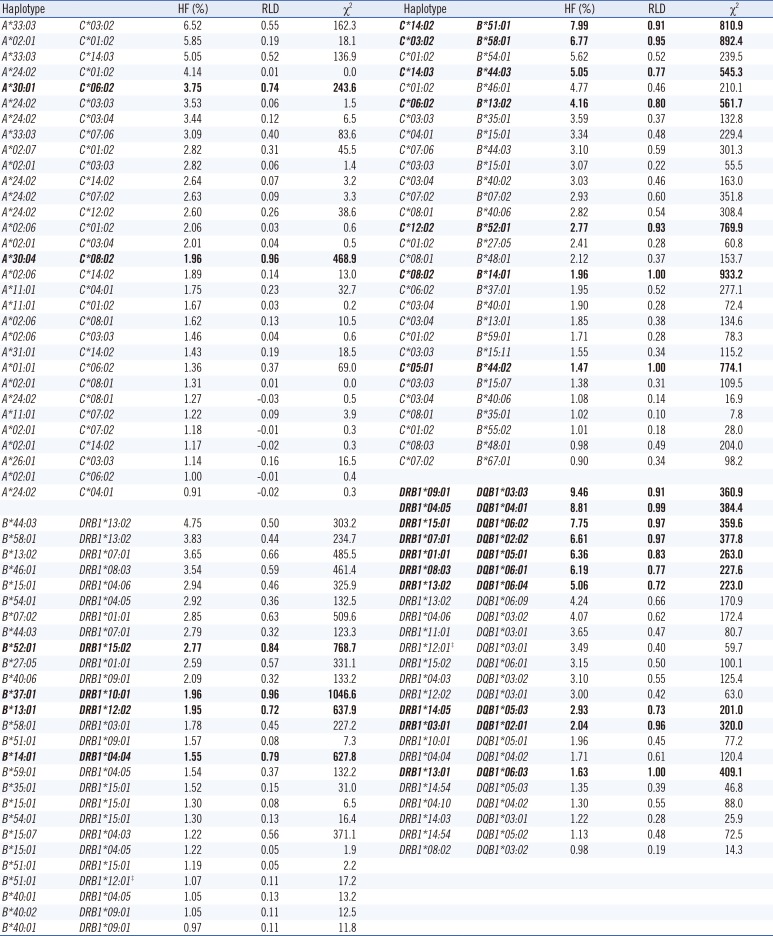
Table 3
HFs (%) of five-locus HLA haplotypes in Koreans† and Japanese
| Haplotypes | HF (%) | ||||||
|---|---|---|---|---|---|---|---|
| A* | C* | B* | DRB1* | DQB1* | Present study | Koreans [4] | Japanese [16] |
| 33:03 | 14:03 | 44:03 | 13:02 | 06:04 | 4.49 | 4.23 | 4.07 |
| 33:03 | 03:02 | 58:01 | 13:02 | 06:09 | 3.34 | 2.99 | 0.19∥ |
| 30:01 | 06:02 | 13:02 | 07:01 | 02:02 | 3.26 | 2.68 | 0.39§ |
| 24:02 | 12:02 | 52:01 | 15:02 | 06:01 | 2.44 | 1.92 | 7.17∥ |
| 33:03 | 07:06 | 44:03 | 07:01 | 02:02 | 2.36 | 2.99‡ | 0.00§ |
| 02:07 | 01:02 | 46:01 | 08:03 | 06:01 | 2.28 | 0.82 | 1.55 |
| 24:02 | 07:02 | 07:02 | 01:01 | 05:01 | 2.16 | 2.89 | 2.71 |
| 02:01 | 01:02 | 27:05 | 01:01 | 05:01 | 1.76 | 1.13 | |
| 30:04 | 08:02 | 14:01 | 04:04 | 04:02 | 1.55 | 1.24 | |
| 11:01 | 04:01 | 15:01 | 04:06 | 03:02 | 1.48 | 1.24 | 1.74 |
| 33:03 | 03:02 | 58:01 | 03:01 | 02:01 | 1.47 | 1.85 | |
| 24:02 | 01:02 | 54:01 | 04:05 | 04:01 | 1.43 | 0.62 | 1.94 |
| 01:01 | 06:02 | 37:01 | 10:01 | 05:01 | 1.13 | 1.03 | 0.39 |
| 24:02 | 01:02 | 59:01 | 04:05 | 04:01 | 1.00 | 1.13 | 0.97 |
| 24:02 | 14:02 | 51:01 | 09:01 | 03:03 | 0.98 | ||
| 02:01 | 03:04 | 13:01 | 12:02 | 03:01 | 0.88 | 0.93 | 0.58 |
| 02:01 | 01:02 | 54:01 | 15:01 | 06:02 | 0.76 | ||
| 11:01 | 01:02 | 54:01 | 04:05 | 04:01 | 0.73 | 0.93 | 1.16 |
| 24:02 | 03:03 | 15:07 | 04:03 | 03:02 | 0.73 | 0.19 | |
| 02:06 | 08:01 | 40:06 | 09:01 | 03:03 | 0.65 | ||
| 03:01 | 05:01 | 44:02 | 13:01 | 06:03 | 0.65 | 0.39 | |
| 24:02 | 03:04 | 13:01 | 12:02 | 03:01 | 0.59 | ||
| 24:02 | 03:04 | 40:02 | 09:01 | 03:03 | 0.57 | 0.78 | |
| 33:03 | 03:02 | 58:01 | 15:01 | 06:02 | 0.57 | ||
Table 4
AFs (%) of HLA-C in Koreans, Japanese, Chinese, and Caucasians
| Serology | HLA-C† |
Koreans‡ (N=613) |
Japanese [17] (N=371) |
Chinese [18] (N=264) |
Caucasians [19] (N=558) |
|---|---|---|---|---|---|
| 1 | *01:02 | 17.8 | 14.8 | 17.4 | 3.7** |
| *01:03 | 0.1 | 0.4 | |||
| *01:04 | 0.1 | ||||
| 2 | *02:02 | 0.5 | 5.8** | ||
| 3 (10) | *03:02 | 7.4 | 0.4** | 8.9 | 0.4** |
| 3 (9) | *03:03 | 11.8 | 12.1 | 3.8** | 5.5¶ |
| 3 (10) | *03:04 | 9.0 | 13.7 | 11.2 | 6.2 |
| 4 | *04:01 | 4.9 | 4.6 | 4.5 | 13.0** |
| 5 | *05:01 | 1.5 | 0.4 | 5.6¶ | |
| 6 | *06:02 | 6.4 | 1.6∥ | 2.8 | 9.8 |
| 7 | *07:01 | 0.2 | 1.7§ | 16.1§,** | |
| *07:02 | 7.3 | 14.6¶ | 21.0** | 11.6 | |
| *07:04 | 0.9 | 0.9 | 0.9 | 2.0 | |
| *07:06 | 3.2 | 0.0** | 0.0∥ | ||
| 8 | *08:01 | 6.9 | 7.4 | 13.3 | 0.2** |
| *08:02 | 2.0 | 0.2 | 3.1 | ||
| *08:03 | 1.0 | 2.0 | 0.1 | ||
| Blank | *12:02 | 3.1 | 10.5** | 1.7 | 0.8 |
| *12:03 | 0.3 | 1.7 | 7.0** | ||
| *14:02 | 8.0 | 4.9 | 3.2 | 1.6** | |
| *14:03 | 5.1 | 8.9 | 0.2∥ | 0.0** | |
| *15:02 | 2.2 | 2.7 | 3.8 | 1.9 | |
| *15:05 | 0.4 | 1.1 | 0.5 | ||
| *16:01 | 2.8** | ||||
| *16:02 | 0.1 | 0.4 |
Table 5
AFs (%) of HLA-DQB1 in Korean, Japanese, Chinese, and Caucasian populations
| Serology | DQB1† |
Korean‡ (N=613) |
Japanese [17] (N=371) |
Chinese [18] (N=264) |
Caucasian [20] (N=250) |
|---|---|---|---|---|---|
| 2 | *02:01 | 2.1 | 0.3§,** | 12.1§ | 26.6§,** |
| *02:02 | 6.7 | ||||
| 3 (7) | *03:01 | 14.0 | 11.3 | 24.2¶ | 15.4 |
| 3 (8) | *03:02 | 9.6 | 10.8 | 7.0 | 10.4 |
| 3 (9) | *03:03 | 11.2 | 13.5 | 15.0 | 6.8 |
| 4 | *04:01 | 8.8 | 11.5 | 4.0 | 0.0** |
| *04:02 | 3.9 | 3.9 | 0.8 | 0.8 | |
| 5 | *05:01 | 9.0 | 7.5 | 3.4 | 0.2** |
| *05:02 | 2.1 | 3.5 | 11.0** | 0.8 | |
| *05:03 | 4.5 | 2.7 | 3.0 | 2.6 | |
| 6 | *06:01 | 9.4 | 18.3** | 11.2 | 0.2** |
| *06:02 | 7.8 | 8.2 | 3.8 | 19.6** | |
| *06:03 | 1.6 | 0.7 | 0.9 | 5.0 | |
| *06:04 | 5.1 | 7.3 | 0.8∥ | 2.0 | |
| *06:09 | 4.2 | 0.4¶ | 1.9 | 0.2∥ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download