Abstract
Background
FcRγ-deficient natural killer (NK) cells (g-NK cells) have been associated with cytomegalovirus (CMV) infection. However, the frequency of g-NK cells in a CMV-endemic area (i.e., Korea) has not yet been studied. We examined the frequency of g-NK cells and expression of CD57 on NK cells in cord blood (CB) and adult blood (AB).
Methods
Of the 24 AB samples collected, 95.8% (23/24) were CMV IgG+/IgM-, while 100% of the 13 healthy CB samples were CMV IgG+/IgM-. We performed whole-blood flow cytometry assays to analyze intracellular FcRγ and CD3ζ expression of CD3-/CD56dim NK cells from 13 CB and 24 AB samples, and surface CD57 expression on CD3-/CD56dim/CD16+ NK cells from 13 CB and 19 AB samples.
Natural killer (NK) cells are innate immune cells that are an integral part of the immune response to certain microbial infections and tumors. In particular, NK cells play a crucial role in the control of herpes virus infection, such as cytomegalovirus (CMV) [1]. NK cell function is regulated by the balance of signals from activating receptors (i.e., NKG2C, NKG2D, NKp30, NKp44, NKp46, CD16), which recognize ligands on tumors and virus-infected cells, and inhibitory receptors (i.e., killer cell immunoglobulin like receptor, NKG2A) that are specific for major histocompatibility complex class I molecules [1,2,3].
For the stimulatory signals from NK cell-activating receptors (i.e., CD16, NKp46, NKp30), transmembrane signaling adaptors for signal transduction are required. NK cells express immunoreceptor tyrosine-based activation motif (ITAM)-bearing adaptor proteins (i.e., FcRγ and CD3ζ), which transmit biochemical signals through ITAMs [2,3,4]. Recently, Hwang et al. [4] identified a distinct subset of human NK cells that are deficient in FcRγ, but express normal levels of CD3ζ, called FcRγ-deficient NK cells (g-NK cells). g-NK cells were readily detectable in 32.0% (39/122) of healthy blood donors in the United States and were confined to the CD56dim population. A subsequent study by the same group also reported that the presence of g-NK cells is strongly associated with previous exposure to CMV and that g-NK cells express significantly higher CD57 and NKG2C levels, but lower NKG2A levels, than conventional NK cells [5].
NK cells expand in response to chronic infections, particularly human CMV. CMV drives expansion of NKG2C+ NK cells, which preferentially acquire CD57 [6]. Therefore, CD57 can be considered a CMV infection-associated marker of NK cells and can be used to determine the association between g-NK cells and prior CMV infections [5].
CMV infection in the general population is asymptomatic; however, the infection mostly establishes life-long latent infection and can be reactivated when hosts become immunocompromised. CMV is found in all geographic locations, socioeconomic groups, and ages. Between 36.3% and 90.8% of adults in the United States are CMV-seropositive [7]. Interestingly, some countries show extremely high CMV-seroprevalence, with that in Korea estimated to be 96% (552/575 individuals) [8,9,10].
The frequency of g-NK cells has been studied only in the US population. However, the frequency of g-NK cells in a CMV-endemic area (i.e., Korea) has not yet been investigated. We hypothesized that the CMV-endemic Korean population has a high frequency of g-NK cells, and attempted to determine the frequency of g-NK cells in both CMV-seropositive and CMV-seronegative individuals. Since CMV IgG-negative samples from healthy adult donors in CMV-endemic Korea are rare, we decided to analyze g-NK cells in umbilical cord blood (CB), which is considered to be CMV-naïve [6].
Here, we examined the frequency of g-NK cells in CB and adult blood (AB) samples from the Korean population. We also investigated the expression of CD57 on NK cells in AB and CB.
All study samples were obtained following acquisition of written informed consent from the study participants or the mothers of infants in the case of CB samples, in accordance with the Declaration of Helsinki. This research protocol was reviewed and approved by the institutional review board of Chonnam National University Hwasun Hospital (CNUHH) (Permit Number: 2012-126).
All adults enrolled in this study were healthy and free of HIV, hepatitis B virus, and hepatitis C virus infections. EDTA blood samples from 37 subjects (24 AB and 13 CB [collected at birth from full-term neonates]) left over from ABO/RhD blood typing or complete blood count analysis were used for the study of g-NK cells, and leftover serum or plasma were used for measurement of anti-CMV IgG and IgM. Among these samples, 19 AB and 13 CB samples were used for the assessment of CD57+ NK cells. All samples were obtained from CNUHH and Chonnam National University Hospital.
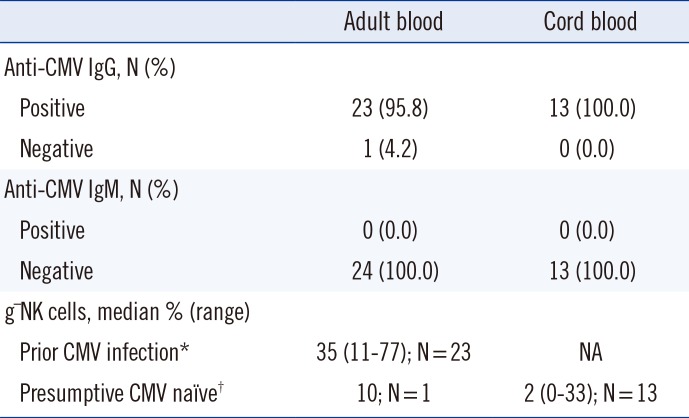
Samples were processed within 24 hr of collection. The presence of anti-CMV IgG and IgM antibodies in the sera of 24 Korean healthy adult donors was determined by chemiluminescent microparticle immunoassay (CMIA) (Architect; Abbott Laboratories, Abbott Park, IL, USA). Of the AB samples, 95.8% (23/24) were CMV IgG+/IgM- and 4.2% (1/24) were CMV IgG-/IgM-, whereas 100.0% (13/13) of the CB samples were CMV IgG+/IgM- (Table 1). Although all CB samples were CMV IgG+, these CB samples were regarded as CMV-negative, because anti-CMV IgG antibodies can cross the placenta. Naitou et al. [11] reported that qualitative nested PCR for CMV DNA was negative in 40 CB plasma samples. This report supports the assumption that CB is a CMV-naïve sample. In addition, Foley et al. [6] also used CB in their study to avoid confounding effects from adult donors who may have encountered CMV previously. On the basis of their findings, the CB samples from 13 healthy neonates used in this study were regarded as CMV-negative controls.
Signaling adaptors (FcRγ and CD3ζ) were detected by using a modification of the previously described method [4]. Instead of using peripheral blood mononuclear cells (PBMCs), whole blood samples (50 µL) were stained for flow cytometry analysis by using phycoerythrin (PE)-conjugated anti-CD3 (clone UCHT1; BD Biosciences, San Jose, CA, USA) and PE-conjugated Cy5-anti-CD56 antibodies (clone B159, BD Biosciences). After surface labeling, the cells were washed, fixed, and permeabilized (IntraPrep kit, Beckman Coulter; Fullerton, CA, USA), according to the manufacturer's instructions. For detection of intracellular FcRγ and CD3ζ expression, fixed and permeabilized cells were stained with fluorescein isothiocyanate (FITC)-anti-FcεRIγ (FcRγ) (Millipore, Temecula, CA, USA) or FITC-anti-CD247 (CD3ζ) (Biolegend, San Diego, CA, USA) antibodies. Samples were acquired with FACSCalibur system (Becton Dickinson, San Jose, CA, USA), and the resulting data were analyzed by using CellQuest software (Becton Dickinson). On the basis of these two signal adaptors' intracellular expression of CD3-/CD56dim NK cells, a distinct subset of human NK cells were identified as g-NK cells; these cells are deficient for FcRγ expression, but express normal levels of CD3ζ. We used 10% as an arbitrary cut-off value to define the presence or absence of g-NK cells for this study.
Whole blood from AB and CB samples was stained with the following antibodies: peridinin chlorophyll protein complex (PerCP)-anti-CD45 (clone 2D1; BD Biosciences), Pacific blue-anti-CD3 (UCHT1, Beckman Coulter), allophycocyanin (APC)-anti-CD56 (clone N901, Beckman Coulter), CD16-PC7 (clone 3G8, Beckman Coulter), and FITC-anti-CD57 (clone HNK-1; BD Biosciences). After incubation, the red blood cells in whole blood samples were lysed as described above. Samples were acquired with NAVIOS flow cytometer (Beckman Coulter). During sample acquisition, low side-scatter (SS) and bright CD45 staining were used to set an electronic gate around the lymphocyte population. The expression of CD57 on NK cells from the CD3-/CD56dim/CD16+gates was analyzed by using Kaluza software (Beckman Coulter).
We determined the frequency of g-NK cells in the CD3-/CD56dim NK cell population. Only one AB sample showed 9.8% g-NK cells, and was thus designated as g-NK cell-negative, according to our arbitrarily chosen cut-off value of 10%. In the remaining AB samples, the proportion of g-NK cells ranged from 11% to as high as 77% (median 35%) (Fig. 1A, B). The one AB donor who had 9.8% g-NK cells was CMV IgG-/IgM-.
We then analyzed the frequency of g-NK cells in the 13 CB samples. Among the 13 CB samples (all samples were anti-CMV IgG+/IgM-, with no clinical evidence of congenital CMV infection), only one sample was designated as g-NK cell-positive, as it showed 33% of g-NK cells in the CD3-/CD56dim NK cell pool. The proportion of g-NK cells in CB samples was significantly lower than that in AB samples (P<0.001; Fig. 1C).
We gated CD45bright/SSClow/CD3-/CD56dim/CD16+ NK cells from 19 AB and 13 CB samples and analyzed the expression of CD57 (Fig. 2A). When CD57 positivity was defined as at least 10% of the CD3-/CD56dim/CD16+ NK cell pool, we could detect CD57+ NK cells in all 19 AB samples tested, with positivity varying from 50.5% to 82.0%. In contrast, less than 10% of these NK cells were detected in all 13 CB samples tested (Fig. 2B).
In the present study, among the 24 AB samples, 95.8% (23/24) were CMV IgG+/IgM-, while 100% of the 13 healthy CB samples were CMV IgG+/IgM-. Studies from other CMV-endemic areas, such as Africa and Asia, also demonstrated a high maternal CMV-seroprevalence (90-100%) [12], consistent with our results. In this study, whole blood was used rather than PBMCs for analysis of g-NK cells and CD57+ NK cells. Single platform flow cytometry with a lyse-no-wash procedure was used to analyze AB and CB samples to overcome the technical difficulties associated with limited CB volumes. Compared with the density gradient separation method for PBMCs isolation, this method reduces loss of any particular lymphocyte subclass because sample manipulation is minimized [13]. For a more clear-cut discrimination between g-NK cells and conventional NK cells, an arbitrary cut-off of 10% was chosen, rather than the 3% cut-off used by Hwang et al. [4].
The frequency of g-NK cells in AB from individuals with prior CMV infection and that in CMV-naïve CB were determined. All CMV-seropositive AB samples contained g-NK cells (23/23), and the proportion of g-NK cells in the CD3-/CD56dim NK cell pool was 35.0% (range, 11-77%). Our results are consistent with a previous report that prior CMV infection is associated with a high frequency of g-NK cells [5]. In addition to the high frequency of g-NK cells, we also found that the proportions of g-NK cells among CD3-/CD56dim NK cells were relatively high compared with those found in healthy US adults [4]. In contrast with CMV, it has been reported that infection with two common herpes viruses (HSV-1 and HSV-2) was not associated with a high frequency of g-NK cells [5].
Recently, CMV has been reported to induce the expansion of CD94/NKG2C+ NK cells in healthy donors as well as in HIV- or hantavirus-infected patients and leukemia patients. The percentage of CD94/NKG2C+ NK cells remains elevated even after therapeutic intervention and in asymptomatic CMV+ donors who likely contracted the virus during childhood [14,15,16,17,18]. Another study found that NKG2C+ NK cells proliferated and acquired CD57 during acute human CMV infection in solid-organ transplant recipients [19]. Furthermore, CD57+NKG2C+ NK cells can be detected in CMV+ healthy adults several years after the primary infection.
In the present study, we examined the expression of CD57 on NK cells in CMV-endemic Korean AB and CB samples. The proportion of CD57+ NK cells in the CD3-/CD56dim/CD16+ cell population in AB was significantly higher than that in CB (P<0.001; Fig. 2B). This finding is consistent with a previous study showing that CD56dim NK cells derived from CB almost completely lacked surface expression of CD57 [20]. However, a significant proportion of CD57+ NK cells in the CD56dim/CD16+ cell subset was also reported in CMV-seronegative young donors [21]. These findings suggest that the different proportions of CD57+ NK cells in AB and CB are associated not only with CMV exposure but also with other factors, such as the immaturity of CB.
We present for the first time a comparative analysis of g-NK cells in AB and CB in the CMV-endemic Korean population. Compared with AB from the US population, AB from the CMV-endemic Korean population had a high frequency of g-NK cells and CD57+ NK cells, whereas CB samples had a very low frequency of g-NK cells and CD57+ NK cells.
Acknowledgments
The authors thank Mr. Yoohyun Kim for his excellent technical assistance. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2010-0023757) and by a grant (CRI13047-21) from Chonnam National University Hospital Research Institute of Clinical Medicine.
References
1. Muntasell A, Vilches C, Angulo A, López-Botet M. Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host pathogen interaction. Eur J Immunol. 2013; 43:1133–1141. PMID: 23552990.
2. Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003; 15:308–314. PMID: 12787756.

3. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008; 9:495–502. PMID: 18425106.

4. Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T, et al. Identification of human NK cells that are deficient for signaling adaptor FcRɣ and specialized for antibody-dependent immune functions. Int Immunol. 2012; 24:793–802. PMID: 22962434.

5. Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRɣ deficiency. J Immunol. 2013; 190:1402–1406. PMID: 23345329.

6. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012; 119:2665–2674. PMID: 22180440.

7. Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006; 43:1143–1151. PMID: 17029132.

8. Lopo S, Vinagre E, Palminha P, Paixao MT, Nogueira P, Freitas MG. Seroprevalence to cytomegalovirus in the Portuguese population, 2002-2003. Euro Surveill. 2011; 16:pii:19896.

9. Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010; 20:202–213. PMID: 20564615.

10. Sohn YM, Park KI, Lee C, Han DG, Lee WY. Congenital cytomegalovirus infection in Korean population with very high prevalence of maternal immunity. J Korean Med Sci. 1992; 7:47–51. PMID: 1329845.

11. Naitou H, Mimaya J, Horikoshi Y, Takashima Y, Amano K, Morita T. Qualitative and quantitative detection of cytomegalovirus DNA in sera by PCR as a clinical marker. Biol Pharm Bull. 1998; 21:1371–1375. PMID: 9881658.

12. Gaytant MA, Steegers EA, Semmekrot BA, Merkus HM, Galama JM. Congenital cytomegalovirus infection: review of the epidemiology and outcome. Obstet Gynecol Surv. 2002; 57:245–256. PMID: 11961482.

13. Tamul KR, Schmitz JL, Kane K, Folds JD. Comparison of the effects of Ficoll-Hypaque separation and whole blood lysis on results of immunophenotypic analysis of blood and bone marrow samples from patients with hematologic malignancies. Clin Diagn Lab Immunol. 1995; 2:337–342. PMID: 7545079.

14. Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2011; 108:14725–14732. PMID: 21825173.
15. Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011; 208:13–21. PMID: 21173105.

16. Marcenaro E, Carlomagno S, Pesce S, Della Chiesa M, Parolini S, Moretta A, et al. NK cells and their receptors during viral infections. Immunotherapy. 2011; 3:1075–1086. PMID: 21913830.

17. Gumá M, Budt M, Sáez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006; 107:3624–3631. PMID: 16384928.
18. Gumá M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, et al. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006; 194:38–41. PMID: 16741880.

19. Della Chiesa M, Marcenaro E, Sivori S, Carlomagno S, Pesce S, Moretta A. Human NK cell response to pathogens. Semin Immunol. 2014; 26:152–160. PMID: 24582551.

20. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010; 116:3853–3864. PMID: 20696944.

21. Campos C, Pera A, Sanchez-Correa B, Alonso C, Lopez-Fernandez I, Morgado S, et al. Effect of age and CMV on NK cell subpopulations. Exp Gerontol. 2014; 54:130–137. PMID: 24440462.

Fig. 1
Identification of FcRγ-deficient human NK cells (g-NK cells) and distribution of g-NK cells in cord blood (CB) and adult blood (AB). (A) Representative flow cytometry plots from one CB and one AB samples. CD3-/CD56dim NK cells in CB express both CD3ζ and FcRγ, whereas NK cells in AB express CD3ζ with low levels of FcRγ. (B) Diagram showing the proportion according to the percentage of g-NK cells among the CD3-/CD56dim NK cells in CB and AB. (C) Comparison of g-NK cells between CB (N=13) and AB (N=24). Horizontal bars represent medians. Mann-Whitney U test was used to compare data between the groups.
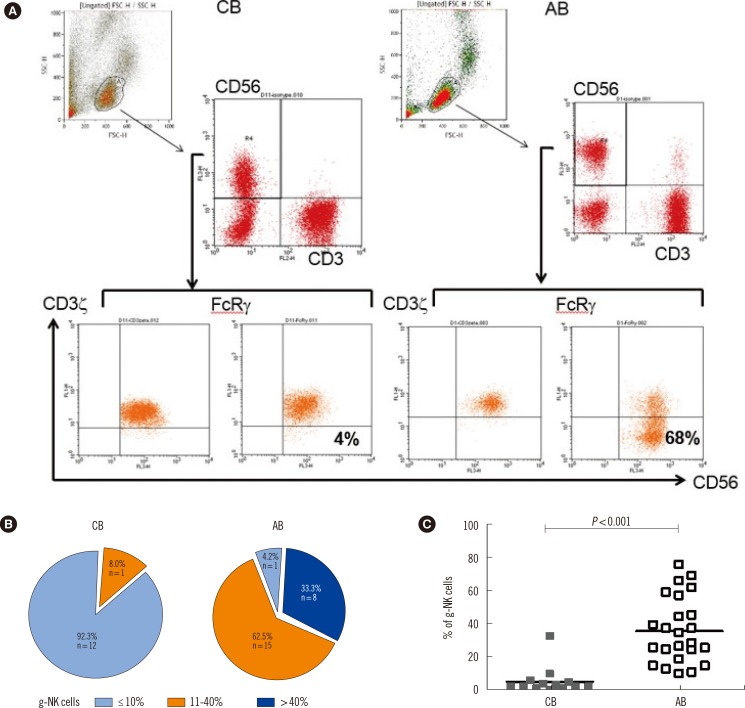
Fig. 2
Distribution of CD57+ cells in cord blood (CB) and adult blood (AB). (A) CD45bright/SSClow/CD3-/CD56dim/CD16+ natural killer (NK) cells from CB (upper panels) and AB (lower panels) were gated and analyzed for CD57 expression. Two representative donors (one CB and one AB) are shown. (B) Comparison of CD57 expression in the CD3-/CD56dim/CD16+ NK cells from CB (N=13) and AB (N=19). Horizontal bars represent medians. Mann-Whitney U test was used to compare data between the groups.
Abbreviation: FITC, fluorescein isothiocyanate.
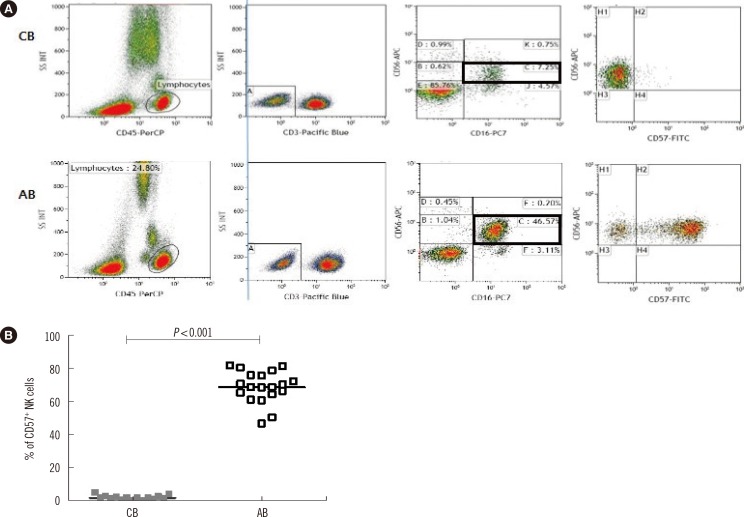
Table 1
Cytomegalovirus antibody status and frequency of g-NK cells in healthy adult and cord blood in the Korean population




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download