INTRODUCTION
METHODS
1. Literature search
2. Inclusion and exclusion criteria
3. Data extraction and assessment
4. Description of studies
Table 1
Summary of the data used in the meta-analysis

Please delete this line. These footnote belong to Fig. 2.
Abbreviations: ARMS, amplification refractory mutation system; AS, allele-specific; RFLP, restriction fragment length polymorphism.
5. Statistical analysis
RESULTS
1. Meta-analysis
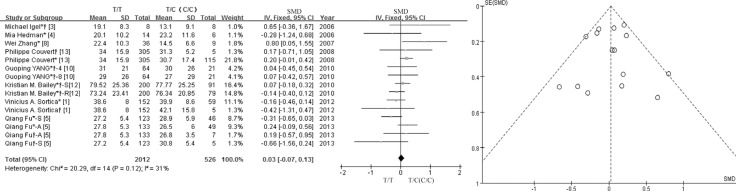 | Fig. 2Forest plot of SMD for the association between the lipid-lowering efficacy of statins and the SLCO1B1 c.521T>C polymorphism, and funnel plots for evaluating publication bias. *TC genotype; †CC genotype; *†TC and CC genotype.Abbreviations: SMD, standard mean difference; CI, confidence interval; df, degrees of freedom; S, simvastatin; A, atorvastatin; R, rosuvastatin; 4, 4 weeks; 8, 8 weeks.
|
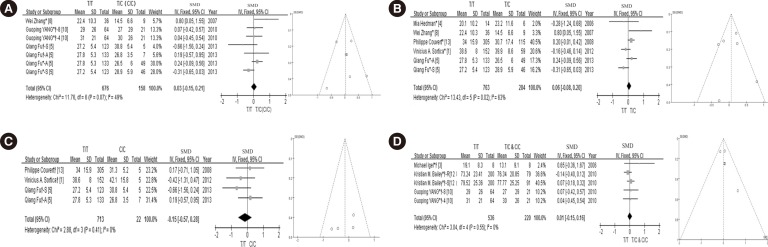 | Fig. 3Forest plots and funnel plots of each subgroup. (A) Forest plots of SMD for the association between the lipid-lowering efficacy of statins and the SLCO1B1 c.521T>C polymorphism of Chinese populations, and funnel plot for evaluating publication bias; (B) Forest plot of SMD for the association between the lipid-lowering efficacy of statins and the SLCO1B1 c.521T>C heterozygous genotype, and funnel plot for evaluating publication bias; (C) Forest plot of SMD for the association between the lipid-lowering efficacy of statins and the SLCO1B1 c.521T>C homozygote genotype, and funnel plotfor evaluating publication bias; (D) Forrest plot of SMD for the association between the lipid-lowering efficacy of statins and the SLCO1B1 c.521T>C variant genotypes (T/C and C/C genotype), and funnel plot for evaluating publication bias. *TC genotype; †CC genotype; *†TC and CC genotype; S: simvastatin; A: atorvastatin; R: rosuvastatin; 4: 4 weeks; 8: 8 weeks.Abbreviations: SMD, standard mean difference; CI, confidence interval; df, degrees of freedom.
|
Table 2
Meta-analysis results of all studies and each subgroup studies

*Analysis of the association between the lipid-lowering efficacy of statins and SLCO1B1 c.521T>C polymorphism; †Analysis of the association between the lipid-lowering efficacy of statins and SLCO1B1 c.521T>C polymorphism in Chinese populations; ‡Analysis of the association between the lipid-lowering efficacy of statins and SLCO1B1 variant heterozygotes (T/C); §Analysis of the association between the lipid-lowering efficacy of statins and SLCO1B1 variant homozygotes (C/C); ∥These studies did not provide the cases of single heterozygote (T/C) or homozygote (C/C).
Abbreviations: SMD, standard mean difference; CI, confidence interval.
Table 3
Main results of meta-analysis for the drug type and treatment length subgroups

*The data by Bailey et al. [12] were excluded. If P>1, there is no heterogeneity in simvastin subgroup.
Abbreviations: SMD, standard mean difference; CI, confidence interval.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download