Abstract
Emerging resistance to trimethoprim/sulfamethoxazole (SXT) poses a serious threat to the treatment of Stenotrophomonas maltophilia infections. We determined the prevalence and molecular characteristics of acquired SXT resistance in recent clinical S. maltophilia isolates obtained from Korea. A total of 252 clinical isolates of S. maltophilia were collected from 10 university hospitals in Korea between 2009 and 2010. Antimicrobial susceptibility was determined by using the CLSI agar dilution method. The sul1, sul2, and sul3 genes, integrons, insertion sequence common region (ISCR) elements, and dfrA genes were detected using PCR. The presence of the sul1 gene and integrons was confirmed through sequence analysis. Among the 32 SXT-resistant isolates, sul1 was detected in 23 isolates (72%), all of which demonstrated high-level resistance (≥64 mg/L) to SXT. The sul1 gene (varying in size and structure) was linked to class 1 integrons in 15 of the 23 isolates (65%) harboring this gene. None of the SXT-susceptible isolates or the SXT-resistant isolates with a minimum inhibitory concentration of 4 and 8 mg/L were positive for sul1. Moreover, the sul2, sul3, and dfrA genes or the ISCR elements were not detected. The sul1 gene may play an important role in the high-level SXT resistance observed in S. maltophilia.
Go to : 
Stenotrophomonas maltophilia, a non-fermentative, gram-negative bacillus frequently found in community or hospital environments, has emerged as an important opportunistic pathogen; it is most commonly associated with respiratory infections in humans. The incidence of hospital-acquired S. maltophilia infections is increasing, and cases of community-acquired S. maltophilia have also been reported [1, 2]. The treatment of S. maltophilia infections is extremely difficult, owing to the intrinsic or acquired resistance to multiple therapeutic drugs, the development of resistance during therapy, and the paucity of clinical therapeutic data [3]. The recommended first-line agent in the treatment of S. maltophilia is trimethoprim/sulfamethoxazole (SXT), because of low incidence of resistance [1]. However, the treatment of S. maltophilia infections has become more problematic, with an increase in the acquired resistance to SXT [4, 5]. The sul genes are known to contribute to the resistance to SXT and have been reported to associate with the class 1 integrons and insertion sequence common region (ISCR) elements [6, 7]. Moreover, it has been reported that the dfrA genes, located in the gene cassettes of the class 1 integrons, lead to a high-level resistance to SXT [8]. However, sul1 has been detected in the SXT-susceptible S. maltophilia isolates [8, 9, 10]. In this study, we analyzed the predominant mechanisms underlying acquired SXT resistance in recent clinical S. maltophilia isolates obtained from Korea.
A total of 252 non-duplicate clinical isolates of S. maltophilia were collected from 10 university hospitals in Korea between 2009 and 2010. The species were identified by using conventional methods and/or the ATB 32GN system (bioMérieux, Marcy l'Etoile, France). The SXT susceptibility was determined by using the CLSI agar dilution method [11]. In order to assess the presence of sul genes and integrons in the isolates, PCR was performed with specific primers (sul1 [Forward, 5'-ATG GTG ACG GTG TTC GGC ATT CTG A-3'; Reverse, 5'-CTA GGC ATG ATC TAA CCC TCG GTC T-3'], sul2 [Forward, 5'-GAA TAA ATC GCT CAT CAT TTT CGG-3'; Reverse, 5'-CGA ATT CTT GCG GTT TCT TTC AGC-3'], sul3 [Forward, 5'-CAT TCT AGA AAA CAG TCG TAG TTC G-3'; Reverse, 5'-CAT CTG CAG CTA ACC TAG GGC TTT GGA-3'], and class 1 integron [Forward, 5'-GCC TGT TCG GTT CGT AAG CT-3'; Reverse, 5'-CGG ATG TTG CGA TTA CTT CG-3']) [7, 12]. The presence of sul1 genes and integrons in the isolates was confirmed by sequencing. PCR analysis was performedby using ISCR-specific primers (ISCR-F, GCG AGT CAA TCG CCC ACT; ISCR-R, CGA CTC TGT GAT GGA TCG AA) to detect ISCR elements in the sul1-positive S. maltophilia isolates [13], and additional PCR analysis was conducted by using certain primer combinations (e.g., sul1-F:ISCR-R or ISCR-F:sul1-R). The presence of dfrA genes in the isolates was determined as per the method by Seputiene et al. [14]. The dfrA genes were grouped according to their sequence similarity, and the primers were designed for analyzing the dfrA genes. Four groups of primers were used to include all previously reported dfrA genes found in S. maltophilia [8].
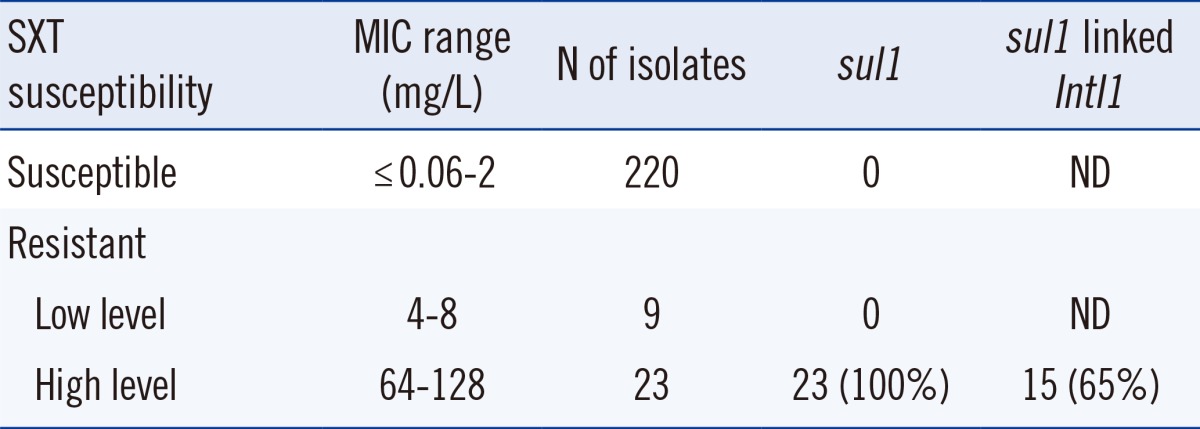
The sul genes were detected in 23 SXT-resistant isolates, but not in nine isolates exhibiting low-level SXT resistance (minimum inhibitory concentration [MIC] 4-8 mg/L). Moreover, sul was not detected in the 220 SXT-susceptible isolates. All the sul genes detected were found to be sul1 genes; the genes sul2 and sul3 were not detected in the isolates. Fifteen of the 23 sul1-positive isolates harbored class 1 integrons (Table 1). The integrons varied in size and structure. Six isolates possessed only intI1 and sul1. All the remaining integrons harbored the qacEΔ1 gene cassettes. One of these integrons also harbored the qac gene. Eight isolates contained one or two aminoglycoside-modifying enzyme genes (four isolates: aac6'-Ib; two isolates: aac6'-31-like; one isolate: aacA7; one isolate: aacA7 and aadA4a). The PCR amplification products of ISCR were not detected, and the linkage of ISCR to the sul1 gene was not observed. No dfrA genes were detected.
In a previous study, the overall resistance rate of the Korean S. maltophilia isolates to SXT was low (4%), while the resistance rate in one hospital was very high (26%) [15]. This result indicates that the empirical selection of SXT for the treatment of S. maltophilia infections in some Korean hospitals may be inadequate for curing this infection completely. Additionally, Song et al. [10] reported that 19 (16%) of the 120 S. maltophilia isolates collected from three university hospitals in a particular region of Korea were found to be resistant to SXT. S. maltophilia isolates are often multidrug-resistant owing to the cumulative effects of intrinsic resistance and acquired resistance through integrons, transposons, and plasmids.
Recently, SXT resistance in S. maltophilia has been reported to be associated with the sul1 gene carried by class 1 integrons [6, 7]. In addition, Toleman et al. [7] demonstrated that the ISCR elements linked to the sul2 genes could mediate SXT resistance in S. maltophilia. Hu et al. [8] hypothesized that the sul1 gene, if combined with the dfrA and sul2 genes, could contribute to SXT resistance. Interestingly, some investigators have reported the detection of the sul1 gene in SXT-susceptible S. maltophilia isolates; however, the incidence of sul1 gene in the SXT-susceptible isolates was lower than that in the SXT-resistant isolates [8, 9, 10].
In this study, of the 32 SXT-resistant S. maltophilia isolates, 23 (72%) harbored the sul1 gene. None of the SXT-susceptible S. maltophilia isolates were found to yield positive sul1 PCR products. These results are similar to those of previous studies [6, 7]. It was not possible to detect the sul1 gene in the nine isolates that demonstrated low-level SXT resistance, although the SXT MIC of the sul1-positive isolates was found to be higher, exhibiting a range of 64-128 mg/L (Table 1). Previous studies reported that S. maltophilia isolates harboring the sul1 gene demonstrated high SXT MICs (>32 mg/L) [7, 8]. Accordingly, it is likely that high-level SXT resistance is associated with a specific and effective mechanism involving sul1.
In contrast to high-level resistance, low-level SXT resistance was not associated with the sul genes. Low-level resistance may result from a much broader variety of biochemical mechanisms than those resulting in high-level resistance. Low antibiotic concentrations can select low-level antibiotic resistant variants, thus producing substantial stress in bacterial populations. This eventually influences the rate of genetic variation and the diversity of adaptive responses. The emergence of low-level resistance should be considered a warning signal, a hallmark of a possible evolutionary trend towards high-level, clinical resistance [16]. Further studies are needed in order to understand the mechanisms and significance of low-level SXT resistance in S. maltophilia.
Usually, the class 1 integrons harbor the sul1 gene at the 3' end [6, 7]. However, there are several reports on sul1-positive isolates not being associated with class 1 integrons [8, 9, 10]. In this study, class 1 integrons were not detected in eight high-level SXT-resistant sul1-positive isolates.
The sul2 gene has been reported to contribute to SXT resistance [7, 9]. However, in this study and in another previous Korean study, the sul2 genes were not detected in the SXT-resistant S. maltophilia clinical isolates [10]. The sul3 gene has not been associated with SXT-resistant S. maltophilia [7]. Contrary to the results reported by Tolman et al. [7], the ISCR elements and the sul2 gene were not detected in this study. Consequently, the association of SXT resistance with the ISCR elements could not be confirmed or further elucidated in this study. Previous reports stated that the dfrA genes were identified in the SXT-resistant isolates and that the sul and dfrA genes could synergistically lead to high-level SXT resistance [8]. In the present study, we could not detect any dfrA genes. Whereas the presence of all types of the dfrA gene was not investigated, the primer pairs for the dfrA genes found in previous reports were included [8].
Previous studies reported the presence of the 3'-end of class 1 integrons, a semiconserved segment harboring the qacEΔ1 and sul1 genes, encoding resistance to quaternary ammonium compounds and sulfonamides, respectively [7, 8, 9]. In this study, the qacEΔ1 genes along with the sul1 genes were detected in nine (60%) class 1 integrons; however, six (40%) class 1 integrons harbored only the intI1 and sul1 genes. The gene cassettes within the class 1 integrons included the aminoglycoside resistance genes aac6'-Ib, aac6'-31-like, aacA7, and aacA7/aadA4a.
In summary, the present results indicate that excessive antibiotic usage in clinical settings is selecting SXT-resistant S. maltophilia strains through horizontal gene transfer. Clinical microbiology laboratories need to carefully monitor, using continuous surveillance of SXT resistance rates, the possibility of S. maltophilia acquiring SXT resistance from mobile elements. The sul1 gene may play an important role in the mechanisms of high-level SXT resistance in S. maltophilia. Additionally, the sul1 gene in the SXT-resistant S. maltophilia has been consistently associated with class 1 integrons. Although SXT resistance is uncommon in S. maltophilia, continuous monitoring of resistance trends is necessary to ensure appropriate antimicrobial therapy.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (NRF-2010-0008485).
Go to : 
Notes
Some of this work was presented at the 52nd Annual Meeting of the Korean Society for Laboratory Medicine, Muju, Korea, October 6-7, 2011 (Oral no. O-27).
Go to : 
References
1. Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. 2009; 9:312–323. PMID: 19393961.
2. Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012; 25:2–41. PMID: 22232370.
3. Farrell DJ, Sader HS, Jones RN. Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrob Agents Chemother. 2010; 54:2735–2737. PMID: 20368399.
4. Tan CK, Liaw SJ, Yu CJ, Teng LJ, Hsueh PR. Extensively drug-resistant Stenotrophomonas maltophilia in a tertiary care hospital in Taiwan: microbiologic characteristics, clinical features, and outcomes. Diagn Microbiol Infect Dis. 2008; 60:205–210. PMID: 17950557.
5. Al-Jasser AM. Stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: an increasing problem. Ann Clin Microbiol Antimicrob. 2006; 5:23. PMID: 16978420.
6. Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, et al. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob Agents Chemother. 2004; 48:666–669. PMID: 14742234.
7. Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 2007; 13:559–565. PMID: 17553270.
8. Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, et al. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents. 2011; 37:230–234. PMID: 21296557.
9. Chang LL, Lin HH, Chang CY, Lu PL. Increased incidence of class 1 integrons in trimethoprim/sulfamethoxazole-resistant clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother. 2007; 59:1038–1039. PMID: 17329265.
10. Song JH, Sung JY, Kwon KC, Park JW, Cho HH, Shin SY, et al. Analysis of acquired resistance genes in Stenotrophomonas maltophilia. Korean J Lab Med. 2010; 30:295–300. PMID: 20603591.
11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing.Twenty-first informational supplement, M100-S21. Wanye, PA: Clinical and Laboratory Standards Institute;2011.
12. Perreten V. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother. 2003; 47:1169–1172. PMID: 12604565.
13. Bae IK, Lee YN, Lee WG, Lee SH, Jeong SH. Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob Agents Chemother. 2007; 51:3017–3019. PMID: 17517851.
14. Seputiene V, Povilonis J, Ruzauskas M, Pavilonis A, Suziedéliené E. Prevalence of trimethoprim resistance genes in Escherichia coli isolates of human and animal origin in Lithuania. J Med Microbiol. 2010; 59:315–322. PMID: 20007760.
15. Chung HS, Hong SG, Kim YR, Shin KS, Whang DH, Ahn JY, et al. Antimicrobial susceptibility of Stenotrophomonas maltophilia isolates from Korea, and the activity of antimicrobial combinations against the isolates. J Korean Med Sci. 2013; 28:62–66. PMID: 23341713.
16. Baquero F. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat. 2001; 4:93–105. PMID: 11512526.

Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download