Dear Editor
The BCR-ABL1 fusion gene is found in about 25% of adult ALL cases [1, 2, 3, 4, 5, 6]. An e1a3-type BCR-ABL1 fusion transcript is found in about 70% of cases, and major breakpoint transcripts (e13a2, e14a2) are found in about 30% of cases. However, only a few cases of ALL with e1a3 BCR-ABL1 fusion transcript have been reported so far [1, 2, 3, 4, 5]. Here, we report two cases of ALL with an e1a3 fusion transcript.
Patients 1 visited our hospital for work-up of back pain and low extremity paresthesia. At diagnosis, white blood cell (WBC), Hb, and platelet levels were 5.530×109/L, 14.2 g/dL, and 113×109/L, respectively. The proportion of blasts reached up to 2% in the peripheral blood and up to 90% in the bone marrow. Immunophenotyping of the blasts revealed the following expression pattern: CD10+, CD19+, CD13+, cytoplasmic CD22 (cCD22)+, cytoplasmic CD79a (cCD79a)+, HLA-DR+, CD34+, cytoplasmic myeloperoxidase (cMPO)-, and nuclear terminal deoxynucleotidyl transferase (nTdT)+. Chromosome analysis showed the following karyotype:
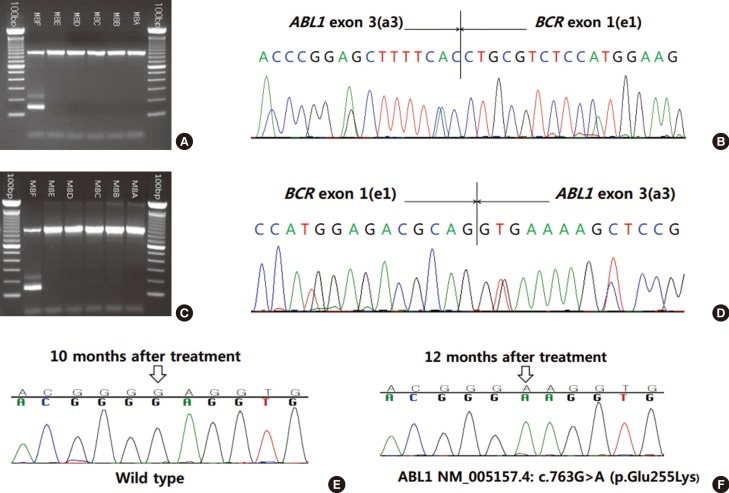
46,XY,t(1;21)(p36.1;q22),t(2;7)(p12;p13),t(9;22)(q34;q11.2)[20]. FISH for BCR-ABL1 (Vysis LSI Dual color dual fusion translocation probe, Abbott Molecular, Des Plaines, IL, USA) showed two fusion signals, suggesting a balanced translocation between BCR and ABL1. Multiplex reverse transcriptase (RT)-PCR using Hemavision kit (DNA Technology, Aarhus, Denmark) revealed an e1a3-type BCR-ABL1 fusion transcript, which was confirmed by sequencing analysis (Fig. 1A, B). Quantitative real-time PCR analysis using the Real-Q BCR-ABL1 Quantification Kit (BioSewoom, Seoul, Korea) failed to detect the BCR-ABL1 fusion transcript. The patient was treated with combination chemotherapy, imatinib, and allogeneic bone marrow transplantation. At the time of writing this manuscript, the patient had been in complete remission for 19 months.
Patient 2 visited our hospital for work-up of fever. At diagnosis, WBC, Hb, and platelet levels were 24.36×109/L, 11.0 g/dL, and 257×109/L, respectively. The proportion of blasts reached up to 70% in the peripheral blood and up to 80% in the bone marrow aspirate. Immunophenotyping showed features of B-cell ALL (B-ALL) with ectopic CD13/CD33 expression (CD19+, CD10+, CD20+, cCD79a+, CD34+, nTdT+, CD13+, CD33+, and cMPO-). Chromosome results were as follows:
46,XY,ider(9)(q10)t(9;22)(q34;q11.2),der(22)t(9;22)[13]/ 46,XY [7], and FISH analysis was positive for BCR-ABL1 (Abbott Molecular). Multiplex RT-PCR (DNA Technology) revealed an e1a3-type BCR-ABL1 transcript, which was confirmed by subsequent sequencing analysis (Fig. 1C, D). Quantitative real-time PCR analysis using the Real-Q BCR-ABL1 Quantification Kit (BioSewoom) failed to detect the BCR-ABL1 fusion transcript. The patient was treated with a combination of chemotherapy and imatinib. However, septic shock and pneumonia developed after induction chemotherapy. Complete remission was not achieved, and the patient died of multi-organ failure after 16 months. A BCR-ABL1 gene mutation study was performed twice, and a missense mutation [ABL1 NM_005157.4: c.763G>A (p.Glu255Lys)] was newly detected in the second evaluation (Fig. 1E, F).
As summarized in Table 1, only a small number of B-ALL cases with the e1a3-type BCR-ABL1 fusion transcript have been reported [1, 2, 3, 4, 5]. Although CML with e1a3 has been reported to be associated with a benign clinical course, the clinical significance of B-ALL with e1a3 is still unclear [6]. In our cases, patient 2 did not respond well to the therapeutic regimen or to imatinib, and underwent an unfavorable clinical course. Furthermore, the patient acquired a mutation associated with imatinib resistance.
Rare fusion transcripts, including e1a3, may not be detected by PCR because of their primer-binding sites [7, 8]. The e1a3-type fusion lacks exon 2 of the ABL1 gene and thus cannot be detected by PCR using primers directed toward sequences in exon 2 [8]. For this reason, we failed to detect the transcript using a commercial real-time quantitative PCR kit, which is designed only for e1a2-, b2a2-, and b3a2-type fusion transcripts. To detect ABL1 gene mutations in these patients, we designed specific primers directed at exon 3 of the ABL1 gene. The primers of ABL1 gene mutation analysis are routinely designed to detect major-bcr since it is the most prevalent (>95%) transcript in CML [9]. Therefore, sequencing analysis of the ABL1 gene in rare-type fusion transcripts using primers for major-bcr may fail to amplify target fusion transcripts.
In conclusion, we report two cases of B-ALL with an e1a3-type BCR-ABL1 fusion transcript. The clinical significance of this rare fusion type is unclear owing to its low incidence [2, 5], but accumulated data suggest that the prognosis is not favorable. Even though currently available commercial multiplex RT-PCR or real-time PCR can detect most types of fusion transcripts, clinicians must be aware of the possibility of false negative results in analyses of rare fusion transcripts [8, 10].
References
1. Fujisawa S, Nakamura S, Naito K, Kobayashi M, Ohnishi K. A variant transcript, e1a3, of the minor BCR-ABL fusion gene in acute lymphoblastic leukemia: case report and review of the literature. Int J Hematol. 2008; 87:184–188. PMID: 18253707.

2. Burmeister T, Schwartz S, Taubald A, Jost E, Lipp T, Schneller F, et al. Atypical BCR-ABL mRNA transcripts in adult acute lymphoblastic leukemia. Haematologica. 2007; 92:1699–1702. PMID: 18055996.

3. Soekarman D, van Denderen J, Hoefsloot L, Moret M, Meeuwsen T, van Baal J, et al. A novel variant of the bcr-abl fusion product in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia. 1990; 4:397–403. PMID: 2193202.
4. Iwata S, Mizutani S, Nakazawa S, Yata J. Heterogeneity of the breakpoint in the ABL gene in cases with BCR/ABL transcript lacking ABL exon a2. Leukemia. 1994; 8:1696–1702. PMID: 7934165.
5. Wilson GA, Vandenberghe EA, Pollitt RC, Rees DC, Goodeve AC, Peake IR, et al. Are aberrant BCR-ABL transcripts more common than previously thought? Br J Haematol. 2000; 111:1109–1111. PMID: 11167748.

6. Roman J, Jimenez A, Barrios M, Castillejo JA, Maldonado J, Torres A. E1A3 as a unique, naturally occurring BCR-ABL transcript in an indolent case of chronic myeloid leukaemia. Br J Haematol. 2001; 114:635–637. PMID: 11552990.

7. Schultheis B, Wang L, Clark RE, Melo JV. BCR-ABL with an e6a2 fusion in a CML patient diagnosed in blast crisis. Leukemia. 2003; 17:2054–2055. PMID: 14513059.

8. Burmeister T, Reinhardt R. A multiplex PCR for improved detection of typical and atypical BCR-ABL fusion transcripts. Leuk Res. 2008; 32:579–585. PMID: 17928051.

9. Goh HG, Hwang JY, Kim SH, Lee YH, Kim YL, Kim DW. Comprehensive analysis of BCR-ABL transcript types in Korean CML patients using a newly developed multiplex RT-PCR. Transl Res. 2006; 148:249–256. PMID: 17145570.

10. Kim HJ, Oh HJ, Lee JW, Jang PS, Chung NG, Kim M, et al. Utility of a multiplex reverse transcriptase-polymerase chain reaction assay (HemaVision) in the evaluation of genetic abnormalities in Korean children with acute leukemia: a single institution study. Korean J Pediatr. 2013; 56:247–253. PMID: 23807891.

Fig. 1
Multiplex reverse transcriptase (RT)-PCR with sequencing analysis and BCR-ABL1 gene mutation study for imatinib resistance. Multiplex RT-PCR and sequencing analysis revealed the e1a3 BCR-ABL1 fusion transcript in patient 1 (A, B) and patient 2 (C, D). BCR-ABL1 gene mutation study was performed for imatinib resistance in patient 2 (E, F). The mutation was detected 12 months after treatment. The 736th nucleotide (empty arrow) of the ABL1 gene was changed from G to A [NM_005157.4: c.763G>A (p.Glu255Lys)].




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download