Abstract
We report three patients with normal karyotype (NK) ALL, who showed genetic aberrations as determined by high-resolution single nucleotide polymorphism array (SNP-A) analysis at both diagnosis and relapse. We evaluated the clinical relevance of the SNP-A assay for the detection of subtle changes in the size of affected genetic lesions at relapse as well as the prognostic value of the assay. In our patients, application of the SNP-A assay enabled sensitive detection of cryptic changes affecting clinically important genes in NK ALL. Therefore, this assay seems to be more advantageous compared to other conventional methods such as FISH assay, HemaVision (DNA Technology, Denmark), and conventional karyotyping for the detection of an "unstable genotype" at relapse, which may be associated with microscopic clonal evolution and poor prognosis. Further comprehensive studies are required to confirm the issues presented by our case patients in this report.
A genome-wide single nucleotide polymorphism array (SNP-A) assay can detect not only the copy number alterations but also the copy-neutral loss of heterozygosity (CN-LOH) on a chromosomal segment with high sensitivity, as demonstrated in MDS/MPN and NK (normal karyotype) AML [1,2,3,4,5]. However, no study has evaluated SNP-A assay in NK ALL, except for one that reported detection of copy number alterations and CN-LOH by SNP-A in 62% of NK ALL patients as well as smaller deletions compared with those in other leukemia subtypes [6]. In addition, the clinical relevance of unstable genotype (subtle changes in the size of detected alterations at relapse) has not been evaluated. We report three patients with NK ALL who harbored genomic alterations, as detected by a high-resolution SNP-A assay at both diagnosis and relapse. We also assessed the clinical relevance of slight changes in the size of affected genetic lesions at relapse with the maintenance of normal cytogenetics.
A 38-yr-old man was diagnosed as having NK ALL on the basis of the results of negative HemaVision (DNA Technology; Aarhus, Denmark) and metaphase cytogenetics (MC) analysis. He was diagnosed as having B-cell ALL with NK and achieved complete remission (CR) at 10 weeks of treatment. He experienced relapse at 8 weeks after CR and, at relapse, his karyotype was maintained as normal and HemaVision results were also negative.
A genome-wide SNP-A assay was performed by using the Affymetrix Cytogenetics 2.7M Array (Affymetrix; Santa Clara, CA, USA) with bone marrow samples obtained both at diagnosis and relapse. At diagnosis, SNP-A analysis revealed homozygous deletions of 9p21.3 (associated with CDKN2A) and 13q14.2 (associated with RB1) and heterozygous deletions of 6p22.2, 8q22.1, 9p21.3p21.2, 14q11.2, 18q22.2, and 19p13.2 (Fig. 1A, B). FISH analysis for the detection of RB1 deletions showed nuc ish (RB1,13q14)×2[200] (Fig. 1C). An identical assay performed at relapse showed loss of heterozygous deletions in 6 affected lesions along with subtle size changes in deleted lesions of 9p21.3 (1,944-1,752 kb) and 13q14.2 (96-89 kb), indicating the persistence of CDKN2A and RB1 deletion at relapse (Fig. 1D, E). FISH analysis for the detection of RB1 deletions also showed nuc ish (RB1,13q14)×2[200] (Fig. 1F).
A 57-yr-old woman was diagnosed as having NK ALL on the basis of the results of HemaVision (negative) and MC analysis. SNP-A analysis at diagnosis revealed heterozygous interstitial deletions of 3q13, 4q23q24, 5q15q21.3, 5q21.3q22.1, 5q22.3q23.1, 5q34, 6q16.3q21, 7q11.23, and 13q14.2q14.3, including RB1 (Fig. 2A). She achieved CR after 4 weeks of treatment but experienced relapse at 6 months after CR. At relapse, her karyotype was normal and HemaVision results were also negative. Results of SNP-A analysis at relapse showed heterozygous deletions of 3q13, 4q23q24, 5q15q21.3, 5q21.3q22.1, 5q22.3q23.1, 5q34, 6q16.3q21, 7q11.23, and 13q14.2q14.3, including RB1 (Fig. 2B), which is similar to those at diagnosis. However, the deleted genomic lesions of 3q13, 4q23q24, 5q15q21.3, 5q21.3q22.1, 5q34, and 13q14.2q14.3 at relapse were slightly different from those at diagnosis.
A 58-yr-old man was diagnosed as having NK ALL as per the results of HemaVision (negative) and MC analysis. He achieved CR after 4 weeks of chemotherapy but experienced relapse at follow-up 32 months after remission. At relapse, the HemaVision results were also negative. However, SNP-A analysis demonstrated an interstitial 451 kb deletion of 9p21.3, which includes interferon α-1/13 (IFNA13) and micro-RNA 31 (MIR31) in a mosaic pattern, at both diagnosis and relapse (Fig. 2C). Demographic findings of the deleted genetic lesions and the affected genes associated with ALL in the three cases are represented in Table 1.
In Case 1, SNP-A analysis could detect the presence of interstitial microdeletion including both RB1 and CDKN2A, which is not otherwise detectable by MC or FISH analysis. Because the deletions of CDKN2A and RB1 are associated with poor prognosis in ALL [7, 8], this may suggest that SNP-A assay can provide prognostic information in NK ALL. The SNP-A assay could detect subtle size changes in the affected lesions (microscopic clonal evolution) and recovery of small-sized interstitial deletions at relapse, which are also not detectable by MC. Thus, the SNP-A assay is more advantageous in detecting microscopic clonal evolution than other methods. In addition, in Case 2, SNP-A assay could detect subtle changes in the size of the affected genetic lesions at relapse (unstable genotype). We may speculate that unstable genotype may indicate microscopic clonal evolution at relapse. The early relapse in Case 2 patient supports this speculation.
The association between ALL and the loss of IFNA13 and MIR31 is unclear. However, as abnormal 9p is an adverse prognostic factor for B-ALL [9], the interstitial deletion of 9p21.3 detected by SNP-A analysis may contribute to poor prognosis, as demonstrated by early relapse in Case 3.
Our report has some limitations. First, the SNP-A assay at relapse in Case 2 did not show any additional abnormalities and slight size changes in the detected genetic lesions as evidence of clonal evolution have not been demonstrated. Therefore, the presence of unstable genotype should be regarded only as a phenomenon suggesting various events, including clonal evolution. Second, we could neither perform SNP-A analysis using samples with CR or fibroblasts, which is important for the discrimination of acquired somatic events from germline aberrations, nor confirm whether all genetic abnormalities detected in our cases were somatic. Third, we could not perform FISH analysis in RB1 in Case 2 as well as detailed evaluations of the discrepancies between the results of FISH and SNP-A analysis. The discrepancy in the results of FISH and SNP-A analysis regarding the RB1 gene in Case 1 may be explained from different detection sensitivity of both methods. However, more comprehensive analysis is required for comprehending this discrepancy.
In conclusion, our case report demonstrates that a SNP-A assay can allow sensitive detection of cryptic changes affecting clinically important genes. A SNP-A assay may be more advantageous than other methods in detecting unstable genotype at relapse, which may be associated with microscopic clonal evolution and poor prognosis in ALL.
Acknowledgments
This work was supported by the 2013 clinical research grant from Pusan National University Hospital.
References
1. Bullinger L, Krönke J, Schön C, Radtke I, Urlbauer K, Botzenhardt U, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010; 24:438–449. PMID: 20016533.

2. Barresi V, Romano A, Musso N, Capizzi C, Consoli C, Martelli MP, et al. Broad copy neutral-loss of heterozygosity regions and rare recurring copy number abnormalities in normal karyotype-acute myeloid leukemia genomes. Genes Chromosomes Cancer. 2010; 49:1014–1023. PMID: 20725993.

3. Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009; 27:5219–5226. PMID: 19770377.

4. Yi JH, Huh J, Kim HJ, Kim SH, Kim HJ, Kim YK, et al. Adverse prognostic impact of abnormal lesions detected by genome-wide single nucleotide polymorphism array-based karyotyping analysis in acute myeloid leukemia with normal karyotype. J Clin Oncol. 2011; 29:4702–4708. PMID: 22084373.

5. Hahm C, Huh HJ, Mun YC, Seong CM, Chung WS, Huh J. Genomic aberrations of myeloproliferative and myelodysplastic/myeloproliferative neoplasms in chronic phase and during disease progression. Int J Lab Hematol. 2014; 5. 21. doi: 10.1111/ijlh.12257. [Epub ahead of print].

6. Huh J, Jung CW, Kim HJ, Kim YK, Moon JH, Sohn SK, et al. Different characteristics identified by single nucleotide polymorphism array analysis in leukemia suggest the need for different application strategies depending on disease category. Genes Chromosomes Cancer. 2013; 52:44–55. PMID: 23023762.

8. Kim M, Yim SH, Cho NS, Kang SH, Ko DH, Oh B, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009; 195:59–65. PMID: 19837270.

9. Heerema NA. 9p rearrangements in ALL. Atlas Genet Cytogenet Oncol Haematol. 1999; 3:153–154. (http://atlasgeneticsoncology.org/Anomalies/9prearrALLID1156.html).

Fig. 1
High-resolution single nucleotide polymorphism array (HR SNP-A) analysis results for chromosome 9 (A) and 13 (B) performed at diagnosis in Case 1. Deleted lesions are indicated with a red box, and detailed array results in each lesion are provided. The results showed a homozygous 1,944 kb deletion of 9p21.3, including CDKN2A; a 96 kb deletion of 13q14.2, including RB1; and a heterozygous 5,262 kb deletion of 9p21.3p21.2. The FISH analysis for the detection of RB1 deletion at diagnosis showed nuc ish (RB1,13q34)×2[200], which indicates no deletion of RB1(C). Identical HR SNP-A analysis results for chromosome 9 (D) and 13 (E) performed at relapse in Case 1 showed a heterozygous 1,752 kb deletion of 9p21.3, including CDKN2A and an 89 kb deletion of 13q14.2, including RB1. FISH analysis for the detection of RB1 deletion at relapse also showed nuc ish (RB1,13q34)×2[200], which indicates no deletion of RB1(F).
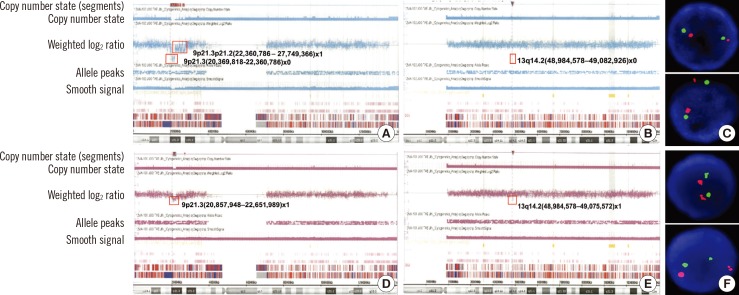
Fig. 2
High-resolution single nucleotide polymorphism array (HR SNP-A) analysis results in whole chromosomes at diagnosis (idiograms of six chromosomes harboring aberrations are provided) in Case 2 (A). The array results showed heterozygous interstitial deletions of 3q13, 4q23q24, 5q15q21.3, 5q21.3q22.1, 5q22.3q23.1, 5q34, 6q16.3q21, 7q11.23, and 13q14.2q14.3, including RB1 gene. Deleted lesions are indicated separately with red arrows, and the detailed array results for each lesion are provided. Identical array results for whole chromosome at relapse in Case 2 (B) showed similar results, but slight size changes in deleted genomic lesions at 3q13, 4q23q24, 5q15q21.3, 5q21.3q22.1, 5q34, and 13q14.2q14.3 were identified at relapse compared to those at diagnosis. Deleted lesions are also indicated separately with red arrows, and detailed array results for each lesion are provided. In addition, HR SNP-A analysis results of chromosome 9 performed at both diagnosis and relapse in Case 3 (C) are provided. The deleted lesion is indicated with a red box, and detailed array result is provided. The array result showed an interstitial 451 kb deletion of 9p21.3, which includes IFNA13 and MIR31 in a mosaic pattern.

Table 1
Characteristics of abnormal lesions detected by high-resolution single nucleotide polymorphism array (SNP-A) assay in 3 cases
*The locations of affected genetic lesions were aligned using the human genome browser-hg19 assembly (http://genome.ucsc.edu/cgi-bin/hgGateway), †The list of affected genes associated with ALL within each genetic lesion was determined from the review of a web-based database (http://atlasgeneticsoncology.org/index.html) and browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
Abbreviations: CDKN2A, cyclin-dependent kinase 2a/p16; RB1, retinoblastoma; TRA, T cell receptor α; TRD, T cell receptor δ.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download