Abstract
Translocations leading to fusions between the immunoglobulin heavy chain gene (IGH) and various partner genes have been reported in B-cell precursor acute lymphoblastic leukemia (B-ALL). However, submicroscopic deletions within IGH in B-ALL have not been rigorously assessed. In this study, we investigated characteristics of IGH submicroscopic deletions, by FISH, in B-ALL with IGH rearrangements. FISH was performed by using commercially available IGH dual-color break-apart rearrangement probes (Abbott/Vysis, Downers Grove, IL, USA; Kreatech, Amsterdam, Netherlands). The study group included seven B-ALL patients with IGH rearrangements, observed by FISH. Among them, two exhibited deletion of the 5' variable region of IGH by FISH. The B-ALL in these two patients included two kinds of abnormal cells; one had an IGH rearrangement without any IGH submicroscopic deletion, while the other had an IGH submicroscopic deletion, which showed that one normal fusion signal and one 3' IGH signal were detected. Thus, submicroscopic deletion of the IGH 5' variable region may have occurred in either the native or rearranged chromosome 14. These findings indicate that B-ALL with IGH rearrangements may be accompanied by submicroscopic deletions of the IGH 5' variable region, which can be detected by FISH. The clinical significance of such deletions is unclear, but the loss of part of the IGH gene in B-ALL warrants further study.
Chromosomal translocations involving the immunoglobulin heavy chain gene (IGH), located on 14q32, are associated with various mature B-cell neoplasms, and result in enhanced expression of the translocation partner genes by physical juxtaposition with enhancers within the IGH locus [1, 2]. Of interest, IGH translocations that give rise to fusions with various partner genes have also been reported in approximately 2-3% of B-cell precursor acute lymphoblastic leukemia (B-ALL) [3, 4, 5]. Among the partner genes, the cytokine receptor-like factor 2 (CRLF2) is the most common, followed by inhibitor of DNA binding 4 (ID4), erythropoietin receptor (EPOR), and the CCAAT enhancer-binding protein (CEBP) family [6]. The misregulated expression of CRLF2, as a consequence of IGH translocation may be associated with a poor prognosis in otherwise "good-risk" patients, suggesting that different IGH chromosomal translocations may constitute subgroups of B-ALL [6, 7].
In addition to IGH rearrangements, IGH submicroscopic deletions are observed in 14-21% of patients with multiple myeloma, and in 13-33% of patients with chronic lymphocytic leukemia [8, 9, 10, 11, 12]. However, IGH submicroscopic deletions in B-ALL have not been thoroughly investigated, and only a few studies have screened for them using Southern blot, FISH, or array comparative genomic hybridization [13, 14, 15]. In this study, we searched for IGH submicroscopic deletions in patients with both B-ALL and IGH rearrangements by FISH using an IGH break-apart probe.
Seven patients with B-ALL and IGH rearrangements by FISH from 2011 to 2013 at the Ewha Womans University School of Medicine and Mokdong Hospital in Seoul, Korea, were enrolled. The diagnoses of B-ALL were made according to the 2008 WHO classification [1], and IGH rearrangements were identified by FISH. Cytogenetic studies were performed on unstimulated 24- and 48-hr cultures of fresh bone marrow aspirates. When possible, at least 20 metaphases per sample were analyzed, and karyotypes were determined according to the International System for Human Cytogenetic Nomenclature (ISCN, 2013) [16]. Interphase FISH was performed by using commercially available probes (Abbott/Vysis; Kreatech). The IGH dual-color break-apart rearrangement probe is a mixture of two fluorescent probes: 5' IGH probe (green) covering the entire IGH variable region, and IGH 3' flanking probe (red) annealing 3' to the constant gene segments of IGH (Fig. 1). The cutoff for IGH break-apart FISH was 3%. At least 200 interphase cells were scored for each probe by two experienced independent examiners. Among seven patients with IGH rearrangements, two had a submicroscopic deletion of the 5' variable region of IGH by FISH (Table 1, cases 1 and 2). They also had BCR/ABL1 and ETV6/RUNX1 rearrangements, respectively. In regards to the IGH rearrangements, two kinds of abnormal cell populations were observed; one had a typical IGH rearrangement without IGH submicroscopic deletion (21% and 18% of interphase cells, in case 1 and 2, respectively) (Fig. 1C-1). The other revealed a submicroscopic deletion of the IGH 5' variable region, resulting in one normal fusion signal and one 3' IGH signal (Fig. 1C-2) (in 22% and 13% of interphase cells, in case 1 and 2, respectively). This finding indicated that an IGH submicroscopic deletion occurred in either the normal or the rearranged chromosome 14 (Fig. 1C-2); we could not distinguish by FISH, which chromosome acquired the deletion. Among the five patients with IGH rearrangements and without IGH submicroscopic deletions, four patients displayed typical IGH rearrangement patterns by FISH (Table 1, cases 3-6; Fig. 1C-1). One patient with hyperdiploidy (Table 1, case 7) exhibited two abnormal cell populations: the first exhibited IGH gain without IGH rearrangement, possibly signifying trisomy 14 in 82% of interphase cells (Fig. 1D-1), and the other, consisting of 10% of interphase cells, showed both IGH gain and IGH rearrangement (Fig. 1D-2).
The biologic significance of these IGH submicroscopic deletions in B-ALL remains to be determined. A previous study using FISH revealed that 6% of pediatric B-ALL patients had IGH submicroscopic deletions without IGH rearrangements, in more than 50% of interphase cells [13]. In the current study, two patients out of seven presented with IGH submicroscopic deletions in 13-22% of their interphase cells (cases 1 and 2); however, we could not distinguish whether IGH submicroscopic deletions developed within normal cells without IGH rearrangements, or in abnormal cells with IGH rearrangements. In most cases of mature B cell neoplasm, submicroscopic deletions of the IGH variable regions occur in the normal chromosome 14, and only rarely in an abnormal chromosome 14 that is involved in an IGH rearrangement [8]. Therefore, some studies suggest that submicroscopic deletions of the IGH variable regions may be a result of the DNA loss that accompanies somatic V(D)J recombination, and may not have any oncogenic role in mature B cell neoplasm [8, 10]. In contrast, one study suggested that relatively large, interstitial 14q deletions in mature B cell neoplasms might activate an unknown oncogene, by operating in a manner similar to translocations that induce fusion of genetic material [17]. This study demonstrated that B-ALL with IGH rearrangements might be accompanied by submicroscopic deletions of IGH 5' variable regions, which can be identified by FISH. A limitation of this study is that the number of patients was relatively small, and further studies in larger cohorts are needed to investigate the incidence and clinical significance of IGH submicroscopic deletions in B-ALL.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2044138).
References
1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC;2008.
2. Meloni-Ehrig A. The cytogenetics of hematologic neoplasms. In : Gersen SL, Keagle MB, editors. The principles of clinical cytogenetics. 3rd ed. New York: Springer;2013. p. 309–370.
3. Dyer MJ, Akasaka T, Capasso M, Dusanjh P, Lee YF, Karran EL, et al. Immunoglobulin heavy chain locus chromosomal translocations in B-cell precursor acute lymphoblastic leukemia: rare clinical curios or potent genetic drivers? Blood. 2010; 115:1490–1499. PMID: 20042721.

4. Akasaka T, Balasas T, Russell LJ, Sugimoto KJ, Majid A, Walewska R, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007; 109:3451–3461. PMID: 17170124.

5. Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009; 144:147–156. PMID: 19006567.

6. Chapiro E, Radford-Weiss I, Cung HA, Dastugue N, Nadal N, Taviaux S, et al. Chromosomal translocations involving the IGH@ locus in B-cell precursor acute lymphoblastic leukemia: 29 new cases and a review of the literature. Cancer Genet. 2013; 206:162–173. PMID: 23827691.

7. Moorman AV, Schwab C, Ensor HM, Russell LJ, Morrison H, Jones L, et al. IGH@ translocations, CRLF2 deregulation, and microdeletions in adolescents and adults with acute lymphoblastic leukemia. J Clin Oncol. 2012; 30:3100–3108. PMID: 22851563.

8. Wlodarska I, Matthews C, Veyt E, Pospisilova H, Catherwood MA, Poulsen TS, et al. Telomeric IGH losses detectable by fluorescence in situ hybridization in chronic lymphocytic leukemia reflect somatic VH recombination events. J Mol Diagn. 2007; 9:47–54. PMID: 17251335.

9. Trakhtenbrot L, Hardan I, Koren-Michowitz M, Oren S, Yshoev G, Rechavi G, et al. Correlation between losses of IGH or its segments and deletions of 13q14 in t(11;14) (q13;q32) multiple myeloma. Genes Chromosomes Cancer. 2010; 49:17–27. PMID: 19787791.
10. Fink SR, Paternoster SF, Smoley SA, Flynn HC, Geyer SM, Shanafelt TD, et al. Fluorescent-labeled DNA probes applied to novel biological aspects of B-cell chronic lymphocytic leukemia. Leuk Res. 2005; 29:253–262. PMID: 15661260.

11. Hwang Y, Lee JY, Mun YC, Seong CM, Chung WS, Huh J. Various patterns of IgH deletion identified by FISH using combined IgH and IgH/CCND1 probes in multiple myeloma and chronic lymphocytic leukemia. Int J Lab Hematol. 2011; 33:299–304. PMID: 21272268.

12. Quintero-Rivera F, Nooraie F, Rao PN. Frequency of 5' IGH deletions in B-cell chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2009; 190:33–39. PMID: 19264231.
13. Hutspardol S, Pakakasama S, Kanta K, Nuntakarn L, Anurathapan U, Sirachainan N, et al. Interphase-FISH screening for eight common rearrangements in pediatric B-cell precursor acute lymphoblastic leukemia. Int J Lab Hematol. 2013; 35:406–415. PMID: 23190578.

14. Dyer MJ, Heward JM, Zani VJ, Buccheri V, Catovsky D. Unusual deletions within the immunoglobulin heavy-chain locus in acute leukemias. Blood. 1993; 82:865–871. PMID: 8338950.

15. Paulsson K, Heidenblad M, Mörse H, Borg A, Fioretos T, Johansson B. Identification of cryptic aberrations and characterization of translocation breakpoints using array CGH in high hyperdiploid childhood acute lymphoblastic leukemia. Leukemia. 2006; 20:2002–2007. PMID: 16990785.

16. Shaffer LG, Jordan JM, Schmid M, editors. An international System for human cytogenetic nomenclature. Basel: S. Karger;2013.
17. Reindl L, Bacher U, Dicker F, Alpermann T, Kern W, Schnittger S, et al. Biological and clinical characterization of recurrent 14q deletions in CLL and other mature B-cell neoplasms. Br J Haematol. 2010; 151:25–36. PMID: 20649559.

Fig. 1
Representative fluorescence in situ hybridization patterns using IGH break-apart probes (green signal: IGH 5' variable region; red signal: IGH 3' flanking probe that lies 3' to the constant gene segments). (A) Normal pattern (2 fusion signals) (B) IGH rearrangement and deletion of the IGH 5' variable region in the normal chromosome 14 (1 green and 2 red signals) (C-1) IGH rearrangement without IGH submicroscopic deletion (1 fusion, 1 green, and 1 red signal) (C-2) one normal IGH locus and submicroscopic deletion of the IGH 5' variable region (1 fusion and 1 red signal). It could not be determined whether the IGH submicroscopic deletion developed within normal cells or cells with an IGH rearrangement. (D-1) gain of IGH, suggesting trisomy 14 (3 normal fusion signals) (D-2) gain of IGH, accompanied by IGH rearrangement (2 fusion signals, 1 green and 1 red signal).
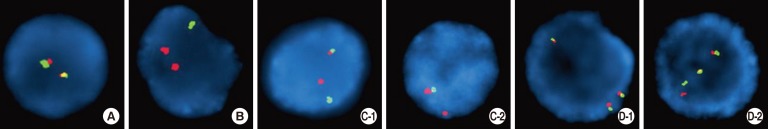
Table 1
Characteristics of IGH rearrangements and IGH deletions in B precursor lymphoblastic leukemia patients
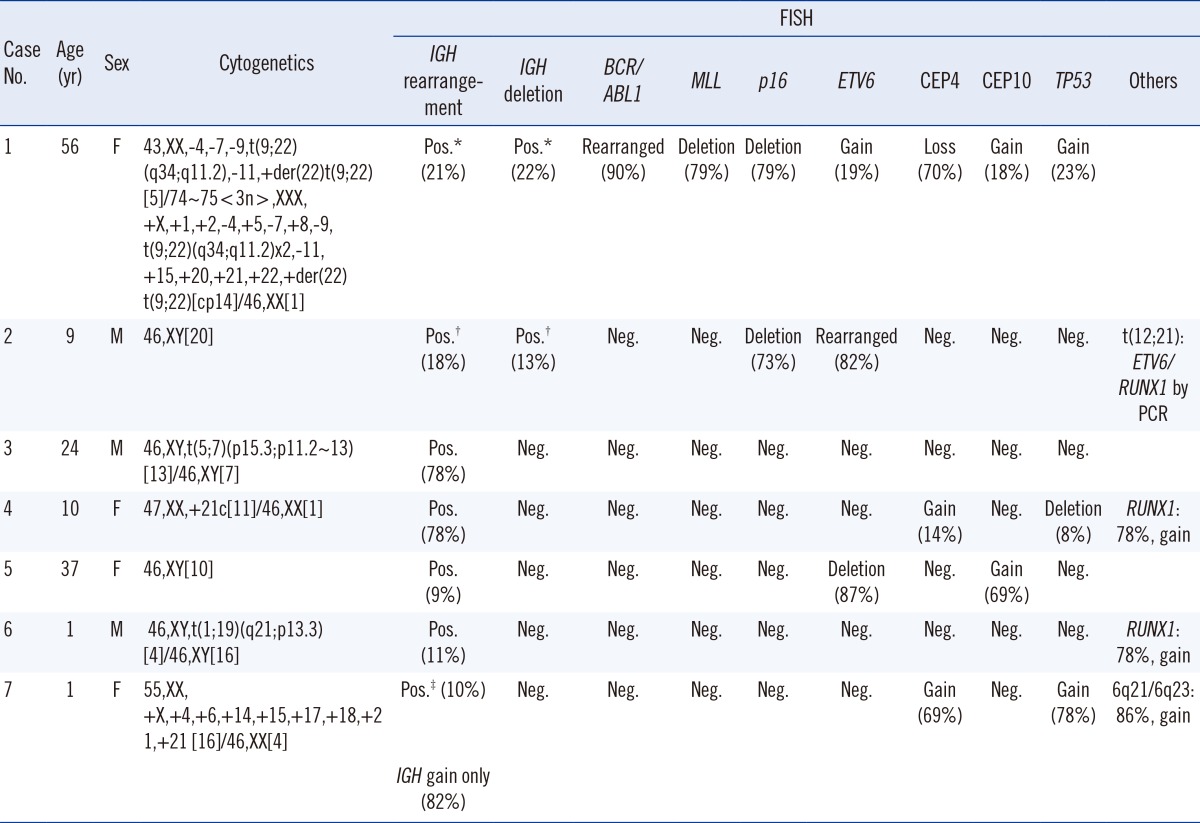
*nuc ish(3'IGHx2,5'IGHx1)(3'IGH con 5'IGHx1)[43/200]/(IGHx2)(3'IGH sep 5'IGHx1)[42/200]; †nuc ish(IGHx2)(3'IGH sep 5'IGHx1)[36/200]/(3'IGHx2,5'IGHx1)(3'IGH con 5'IGHx1)[25/200]; ‡nuc ish(IGHx3)[163/200]/(IGHx3)(3'IGH sep 5'IGHx1)[19/200].
Abbreviations: IGH, immunoglobulin heavy chain gene; CEP, chromosome enumeration probe; Pos., positive; Neg., negative.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download