Measurement of bacterial burden is useful for determining disease severity and transmission risk in tuberculosis (TB). Recently, Xpert MTB/RIF (Cepheid, Sunnyvale, CA, USA) was offered as a new approach for measuring
Mycobacterium tuberculosis (MTB) burden [
1,
2]. The AdvanSure TB/NTM real-time PCR kit (LG Life Science, Seoul, Korea) was developed to detect MTB and non-tuberculous mycobacteria (NTM); it targets the insertion sequence, IS6110 and internal transcribed spacer (ITS) region, respectively. Although this kit showed high sensitivity and specificity [
3-
5], its competency for quantification of bacterial burden has not been studied. In this study, we evaluated the quantitative performance of AdvanSure TB/NTM assay by comparing cycle threshold (Ct) values with 1) acid-fast bacilli (AFB) staining grades and semi-quantitative colony counts on solid medium and 2) time to positivity (TTP) in solid and liquid media. Moreover, we investigated the cutoff Ct value that can best predict smear positivity.
From January to July 2012, we retrospectively analyzed data from 108 patients whose respiratory specimens tested positive for MTB with the AdvanSure TB/NTM kit. Eighty-five specimens were culture positive (69 were grown in both MGIT [Becton Dickinson diagnostic instrument systems, Sparks, MD, USA] and Ogawa media [Asan Pharmaceutical Co., Seoul, Korea] and 16 were grown only in MGIT), and 23 were culture negative. For the culture-positive specimens, we compared Ct values and phenotypical observations. Each patient provided a specimen for real-time PCR and 1-3 specimens for phenotypical testing. This study was approved by the Institutional Review Board of School of Medicine, The Catholic University of Seoul, Korea (KC12SISI0870).
All specimens were liquefied and decontaminated with N-acetyl-L-cysteine-sodium hydroxide (NALC-NaOH). Following concentration, a part of the sediment from each specimen was used for AFB staining; it was cultured on 3% Ogawa medium (Asan Pharmaceutical Co.) and BACTEC MGIT 960 system (Becton Dickinson diagnostic instrument systems). After 4-week culture on Ogawa medium, we counted the number of colonies and graded them according to the Centers for Disease Control and Prevention (CDC) recommended criteria on reporting of colony counts [
6]. As multiple specimens were obtained from each patient, the grades of AFB staining, colony count on Ogawa medium, and the TTP in microbiological culturing were determined as the mean value of multiple specimens.
DNA extract from each specimen was amplified using the AdvanSure TB/NTM real-time PCR kit, following the manufacturer's protocol. According to the manufacturer's instructions, a Ct value of <35 was considered positive for MTB. To investigate the dynamic range of AdvanSure TB/NTM assay, the DNA from MTB strain H37Ra ATCC 35835 was diluted to final concentrations ranging from 108 to 102 CFU/mL (n=5 per dilution) and tested with the AdvanSure TB/NTM assay
Statistical analysis was done using SPSS version 12.0 for Windows (SPSS Inc., Chicago, IL, USA). We compared the Ct values of culture-positive and culture-negative specimens by independent t-test. The strength of correlation between Ct value and grades of AFB staining, solid medium culture, and TTP in microbiological culturing was determined by calculating the Spearman rank correlation coefficient (rs). The correlation between Ct values and solid culture grades was analyzed with Kruskal-Wallis test. We defined all specimens with an AFB staining grade of "trace or above" as stain positive, and ROC was generated with MedCalc version 12.1.4 (MedCalc Software, Mariakerke, Belgium).
The assay identified MTB in all samples containing 10
8 to 10
2 CFU/mL and the log-linear relationship between the Ct value and MTB cells was maintained between 10
8 and 10
2 CFU/mL, confirming the ability of the assay to provide semi-quantitative estimates of input cell number (y=40.617-3.339x, r
2=0.999) (
Fig. 1). Reproducibility was excellent, with a mean coefficient of variance of 1.3% across all dilutions.
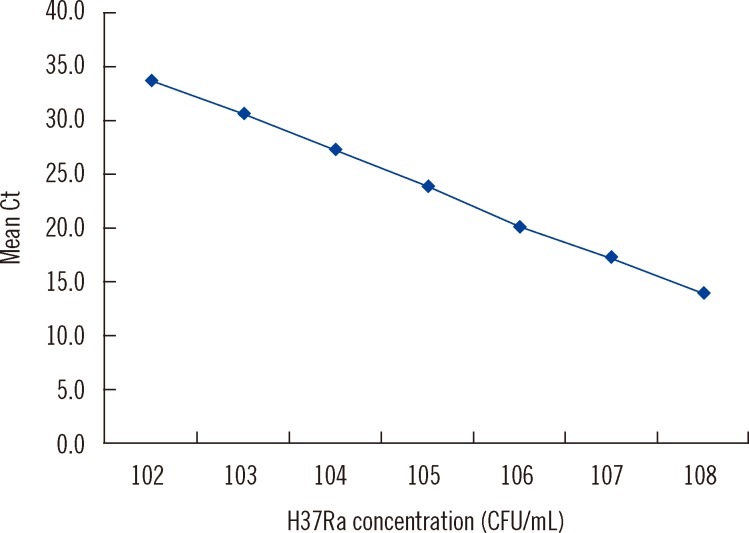 | Fig. 1
Dynamic range test. Log dilution of the DNA extract of MTB strain H37Ra range from 102 to 108 CFU/mL (n=5 per dilution). Cell concentrations fall within the linear range between 102 and 108 CFU/mL (y=40.617-3.339 x, r2=0.999).
Abbreviation: Ct, cycle threshold.

|
The mean Ct values were significantly lower for MTB-culture positive specimens compared to those for MTB culture-negative specimens (28.6 vs. 32.1,
P<0.001). Of the 23 patients with negative results, 18 patients were undergoing treatment for TB, two had history of TB, and the remaining five showed no evidence of active TB. The mean Ct value of positivity on both MGIT and Ogawa media (n=69) was lower than that obtained with MGIT alone (n=16), but the difference was not statistically significant (28.0 [95% CI; 26.9-29.0] vs. 31.4 [95% CI; 29.5-33.3],
P=0.069). The Ct value and AFB staining grade showed a significant reverse correlation (
rs=-0.635,
P<0.01) (
Fig. 2).
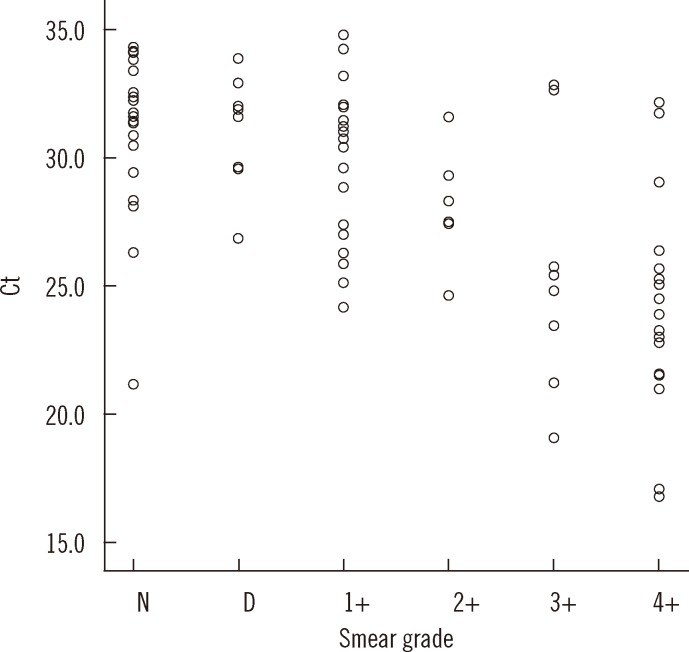 | Fig. 2
Distribution of Ct values according to AFB staining grade. Ct value showed strong reverse correlation with acid-fast bacilli (AFB) staining grade (correlation coefficient, rs=-0.635, P<0.01).
Abbreviations: Ct, cycle threshold; D, doubtful; N, negative.

|
On comparing the culture techniques, the Ct value significantly decreased as the grades of growth on Ogawa medium (n=69) increased (group 1+: 29.0; 2+: 30.0; 3+: 27.1; 4+: 25.5,
P<0.001), but the correlation was weak (
rs=-0.434) (
Fig. 3). The correlation of Ct value with TTP in MGIT (n=85) and Ogawa (n=69) media was 0.468 and 0.365, respectively (
P<0.01) (
Fig. 4).
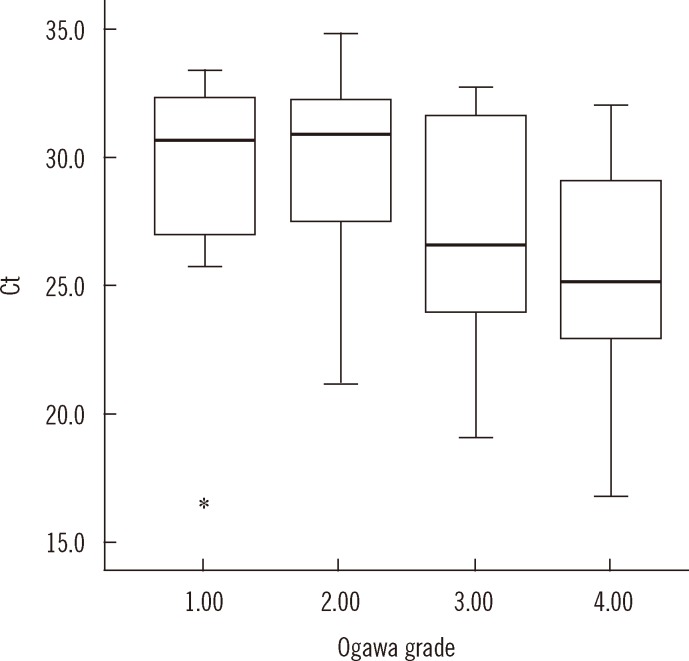 | Fig. 3
Distribution of Ct value with semi-quantitative culture grades on Ogawa medium. The mean Ct value (95% confidence interval [CI]) is 29.0 (25.4-32.5), 30.0 (28.6-31.4), 27.1 (24.6-29.7), and 25.5 (23.6-27.4) in that order. The Ct value tends to decrease with increasing culture grades (P<0.001). The top and bottom borders of the box means 95% CI of the mean Ct. The bars above and below the box means minimum and maximum of the Ct (17.1-33.4, 21.2-34.8, 19.1-32.7, 16.8-32.0, in that order), bold horizontal line in the box is median Ct (30.6, 30.9, 26.6, 25.2, in that order). The real minimum Ct value in grade 1 (17.1) was apart from the mean Ct too far and marked as *on the graph.
Abbreviation: Ct, cycle threshold.

|
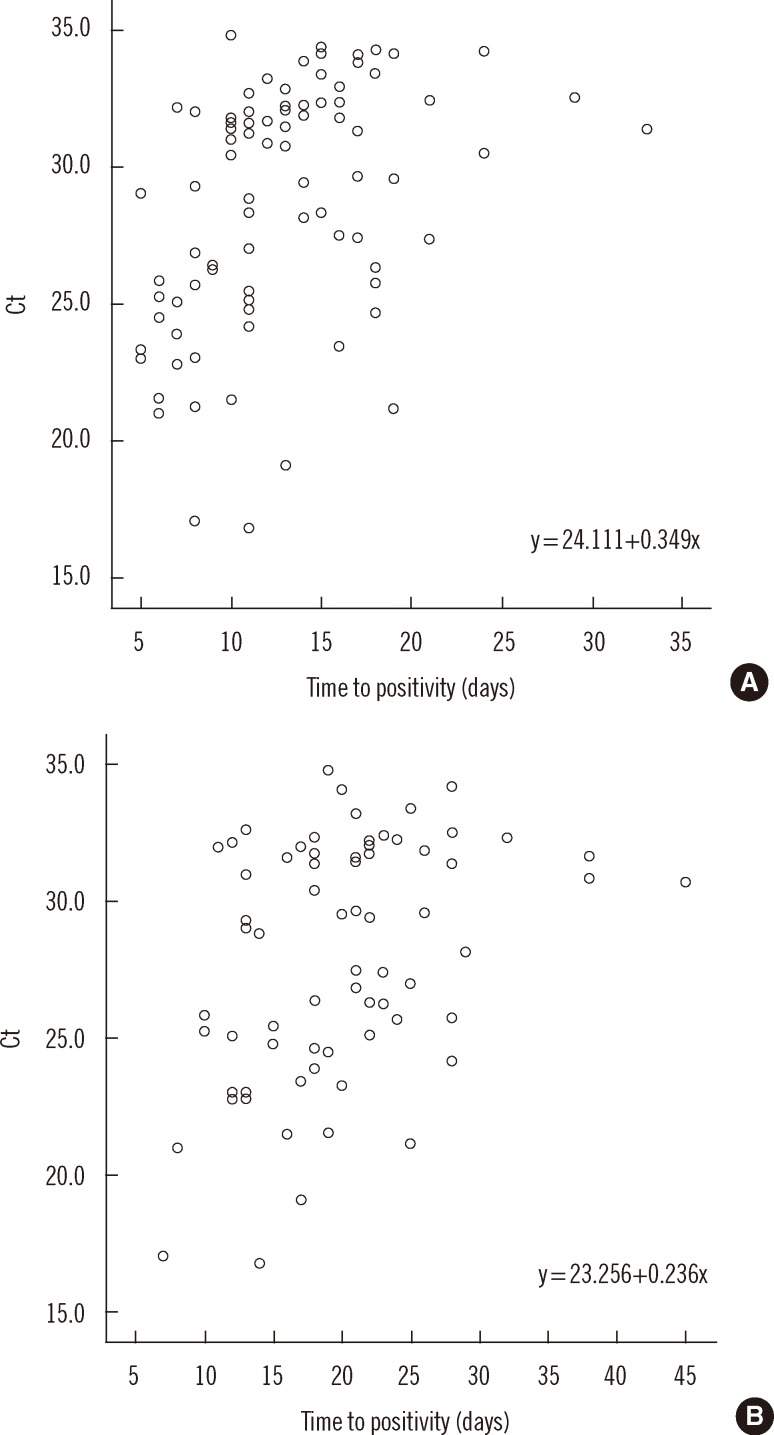 | Fig. 4
Distribution of Ct values according to time to positivity (TTP) in MGIT (A) and in Ogawa (B) media. Ct value showed weak correlation with TTP (correlation coefficient, rs=0.468 and 0.365, respectively, P<0.01).
Abbreviation: Ct, cycle threshold.

|
Between the methods of phenotypical testing, the correlation coefficient between AFB staining and TTP in MGIT and Ogawa media was -0.694 and -0.646, respectively (P<0.01). The correlation coefficient between the semi-quantitative colony grades on Ogawa medium and TTP in MGIT and Ogawa media was -0.611 and -0.737, respectively (P=0.01).
The sensitivity and specificity of various cutoff Ct values for predicting stain positivity (AFB staining grade of "trace or greater") was examined (
Fig. 5). A cutoff Ct value of <33.2 best predicted stain positivity, with a sensitivity of 95.0% (95% CI, 86.1-99.0%) and specificity of 32.0% (95% CI, 14.9-53.5%).
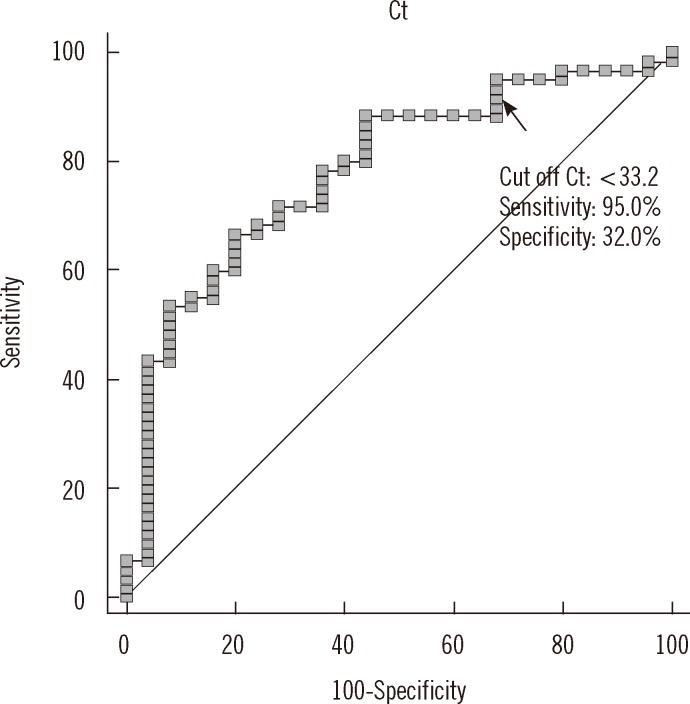 | Fig. 5
ROC curve analysis showing cutoff Ct value for stain positivity. Stain positivity was defined for all concentrated specimens that were graded as "trace or greater." A cutoff Ct value of <33.2 best predicted stain positivity, with a sensitivity of 95.0% (95% CI, 86.1-99.0%), specificity of 32.0% (95% CI, 14.9-53.5%), positive predictive value of 77.0% (95% CI, 65.8-86.0%), and negative predictive value of 72.7% (95% CI, 39.0-94.0%).
Abbreviation: Ct, cycle threshold.

|
A significant reverse correlation was observed between Ct values and AFB staining grades (
rs=-0.635,
P<0.01), and it was comparable to those between staining grades and TTP in MGIT and Ogawa media (
rs=-0.694 and -0.646, respectively). This is in line with a previous study that evaluated the quantitative capabilities of the Xpert MTB/RIF assay where a strong correlation was noted between the Ct value and AFB staining grade (
rs=-0.71) [
1]. However, we found a relatively large variation of Ct value within each smear grade and an overlap of Ct value between adjoining smear grades. This finding is likely because of the operator dependency of smear classification [
7] and because of variation in IS6110 copy number as it varies between 9 and 22 even within the Beijing family strains [
8], which comprise about 78.1% of Korean clinical isolates of MTB [
9]. Although rare in Korea, some MTB strains with no copy of IS6110 were reported [
10].
On comparing with the culture techniques, although the Ct value significantly decreased as Ogawa medium grades increased (
P<0.001), the correlation between Ct value and TTP in MGIT or Ogawa media was weaker (
rs=0.468 and 0.365, respectively) than with the staining grades (
rs=-0.635), and the change in Ct value between culture grades 1+ and 2+ was minor. This finding was also reported for Xpert MTB/RIF, which targets the single-copy rpoB gene; the correlation with AFB staining grade was stronger (
r=-0.77) than with the culture grade (
r=-0.56). There was considerable variation in Xpert's quantitative estimate among sputum samples within the same AFB staining grade [
1].
This weak correlation with culture technique is not surprising because of several reasons: first, variability in the NALC-NaOH decontamination procedure can introduce significant variation in the percentage of viable cells; second, we did not exclude patients undergoing treatment for TB, and MTB DNA persists well beyond time points that render cultures negative, i.e., 1-2 months [
11]. Again, the variation of IS6110 copy number might contribute to the weak correlation.
Of the 23 PCR-positive, culture-negative cases, 18 were explained because they were undergoing treatment for TB or had history of TB. While PCR would detect both viable and dead mycobacteria, microbiological culture will detect only viable mycobacteria. In the remaining five patients, the false-positive reaction could be produced by an unknown gene having sequence similarity with IS6110, or considering the intermediate TB burden in Korea, low numbers of MTB colonized in respiratory tract of patients without active TB could be detected by PCR [
12].
Despite the relatively strong correlation between Ct values and staining grades, it is difficult to select a Ct value that is a sensitive and specific predictor of smear positivity. It can primarily be attributed to low specificity, and we assume that this is due to the difference in the dynamic range, that is, 10
4 to 10
8 CFU/mL for AFB staining [
1] and 10
2 to 10
8 CFU/mL for AdvanSure TB/NTM assay. Considering that smear-negative patients can also be a substantial source of TB [
13], a more sensitive method for early detection of MTB is needed; the Ct value can substitute AFB staining as an indicator of MTB burden. In this study, however, the specificity was too low (32%), in fact a bit lower than that of Xpert MTB/RIF (48%) [
1], and this lower specificity can be due to the variation in IS6110 copy number. Further, in contrast to the study with Xpert MTB/RIF [
1], we used different specimens for molecular and phenotypical testing.
Thus, our preliminary findings suggest the potential use of AdvanSure TB/NTM real-time PCR kit for quantitatively determining bacterial burden, albeit with some enhancements. These findings need to be supported by further studies with larger sample size.






 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download