Abstract
Background
Acinetobacter baumannii is one of the most important pathogens capable of colonization in burn patients, leading to drug-resistant wound infections. This study evaluated the distribution of the AdeABC efflux system genes and their relationship to ciprofloxacin resistance in A. baumannii isolates collected from burn patients.
Methods
A total of 68 A. baumannii clinical strains were isolated from patients hospitalized in Motahari Burns Center in Tehran, Iran. Ciprofloxacin susceptibility was tested by the disk diffusion and agar dilution methods. PCR amplification of the adeRS-adeB drug efflux genes was performed for all resistant and susceptible isolates. To assess the role of the drug efflux pump in ciprofloxacin susceptibility, carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was used as an efflux pump inhibitor (EPI).
Results
Approximately 95.6% of the Acinetobacter isolates were resistant to ciprofloxacin, with minimum inhibitory concentration (MIC) values ranging from 4 to ≥128 µg/mL. The susceptibility of 86.1% of the resistant isolates increased by factors of 2 to 64 in the presence of CCCP. All resistant isolates were positive for the adeRS-adeB genes, and 73.2% of them had mutations in the AdeRS regulatory system.
Conclusions
The results showed that AdeABC genes are common in A. baumannii, which might be associated with ciprofloxacin non-susceptibility, as indicated by the observed linkage to the presence of the genes essential for the activity of the AdeABC, several single mutations occurring in the adeRS regulatory system, and an increase of ciprofloxacin susceptibility in the presence of a CCCP EPI.
Burn injuries are one of the most important health problems in many countries [1, 2]. Organisms associated with nosocomial infections in burn patients include normal flora from exogenous sources in the environment and from healthcare personnel. Currently, Acinetobacter baumannii and Pseudomonas aeruginosa are two of the most common causes of burn wound infections [3, 4]. Of great concern is the spread of A. baumannii strains because of their ability to develop resistance to multiple commonly used antibiotics, including fluoroquinolones. Multidrug resistance is often responsible for the failure of antibiotic therapy [5, 6]. Fluoroquinolones, such as ciprofloxacin (CIP), are very potent antimicrobials that are used as first line antibiotics against A. baumannii infections [7]. Resistance to fluoroquinolones is mediated primarily by spontaneous mutations in their targets, DNA gyrase and topoisomerase IV [7, 8]. A secondary mechanism responsible for fluoroquinolone resistance is reduction in drug accumulation due to overexpression of active efflux pumps [7, 9, 10]. In an energy dependent manner, bacterial drug efflux systems pump out a wide range of antibacterial agents, including antibiotics, biocides, and solvents, without alteration or degradation. In such conditions, the intracellular antibiotic concentration is decreased, and bacteria become less susceptible to the compound [10, 11].
Recently, the role of the AdeABC efflux pump in Acinetobacter drug resistance was described [12, 13]. This efflux pump belongs to the resistance-nodulation-cell division (RND) family and has a three-component structure: AdeA is the membrane fusion protein, AdeB is the multidrug transporter, and AdeC is the outer membrane protein. The adeABC operon is strongly regulated by a two-component system (AdeR-AdeS): AdeS is a sensor kinase and AdeR is a response regulator. Overexpression of the AdeABC efflux pump can be caused either by the point mutations in AdeRS or by the insertion sequence (IS) Aba-1 insertion upstream of the adeABC operon [12, 13, 14]. Single point mutations in adeR (Pro116Leu) and adeS (Thr153Met) are known to be associated with AdeABC overexpression [13], and, subsequently, with resistance to several antibiotics, including aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol, and β-lactams [12, 13]. However, these mutations have not been observed in a small number of clinical isolates with increased levels of expression of AdeABC [15, 16].
Several studies in Iran found increased fluoroquinolone resistance among clinical isolates of A. baumannii and a spread of drug-resistant strains among burn patients in Tehran hospitals. However, the efflux pumps, including those of the RND family that produce multidrug resistance in A. baumannii isolates have not been investigated. In this study, we assessed the association of the AdeABC efflux genes with CIP non-susceptibility in A. baumannii isolates.
Sixty-eight clinical isolates of A. baumannii recovered from patients admitted to the burn unit of Motahari Hospital in Tehran, Iran during the latter part of 2011 were selected for this study. After the burn wound exudates were sampled for clinical specimens, they were examined microbiologically. Bacterial isolates were identified as A. baumannii by using standard biochemical procedures according to the criteria of Bouvet and Grimont [17]. Identifications were confirmed by PCR amplification of the intrinsic blaOXA-51-like gene [18, 19].
Antimicrobial susceptibility tests were performed by using the Kirby-Bauer disk diffusion agar method on Mueller-Hinton (M-H) agar plates (Merck; Darmstadt, Germany) to designate isolates as either ciprofloxacin-resistant A. baumannii (CRAB) or ciprofloxacin-susceptible A. baumannii (CSAB). The minimum inhibitory concentration (MIC) of CIP against CRAB isolates was evaluated by using the agar dilution technique. Both of these methods were performed according tothe CLSI guidelines [20]. Pseudomonas aeruginosa ATCC 27853 was used as the control strain in susceptibility testing.
The presence of one structural (adeB) and two regulatory (adeR and adeS) genes of the AdeABC system were investigated by PCR in CRAB and CSAB isolates. The three pairs of oligonucleotide primers were: adeB primer, adeB-F (5'-TTAACGATAGCGTTGTAACC-3') and adeB-R (5'-TGAGCAGACAATGGAATAGT-3');adeR primer, adeR-F (5'-ACTACGATATTGGCGACATT-3') and adeR-R (5'-GCGTCAGATTAAGCAAGATT-3'); and adeS primer, adeS-F (5'-TTGGTTAGCCACTGTTATCT-3') and adeS-R (5'-AGTGGACGTTAGGTCAAGTT-3'). Genomic DNA was extracted from bacterial colonies by using the Genomic DNA Purification Kit (Fermentase; Vilnius, Lithuania) according to the manufacturer's instructions. Each PCR reaction mixture contained 10 µL of 2× Master Mix (Ampliqon; Odense, Denmark), including 1× PCR buffer, 1.5 mmol/L MgCl2, 0.15 mmol/L dNTP, 1.25 IU TaqDNA polymerase, 0.5 µL of 0.8 µM of each primer, 1 µL of template DNA (0.5 µg), and sterile distilled water up to 25 µL. DNA amplification was carried out in a Mastercycler gradient (Eppendorf; Hamburg, Germany) instrument with initial denaturation at 94℃ for 5 min followed by 30 cycles of amplification (denaturation at 94℃ for 1 min, then annealing at 57℃ for 1 min, and extension at 72℃ for 1 min), ending with a final extension at 72℃ for 5 min. The amplified products were resolved by electrophoresis on 2% agarose gels stained with ethidium bromide. Sequencing of the PCR products of the adeR and adeS genes in 56 CRAB and CSAB isolates with or without active efflux pumps, respectively, was performed by using an ABI 3730XL DNA Analyzer (Applied Biosystem Inc., Forster City, CA, USA). The sequences were compared with GenBank genes by using the BLAST tool available on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/).
A functional efflux system was assessed by measuring the MICs for CIP before and after exposure to the efflux pump inhibitor (EPI), carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma-Aldrich, Dorset, United Kingdom). This compound is an uncoupler of oxidative phosphorylation, which disrupts the proton gradient of the membranes that is required for activity of RND-type pumps [15]. Therefore, addition of CCCP to M-H agar plates leads to increased accumulation of antibiotic and, consequently, reduction of the MIC in isolates that carry active efflux pumps. Briefly, CCCP was added to each of the M-H agar plates containing 0.5-128 µg/mL CIP at a final concentration of 25 µg/mL [21]. The MIC of CIP was determined for all CRAB and CSAB isolates against the A. baumannii ATCC 19606 strain. The plate with CCCP that did not contain antibiotic was used as a control. The effects ofthe EPI were determined by detecting a 4-fold or greater increase in susceptibility after incorporation of CCCP [21].
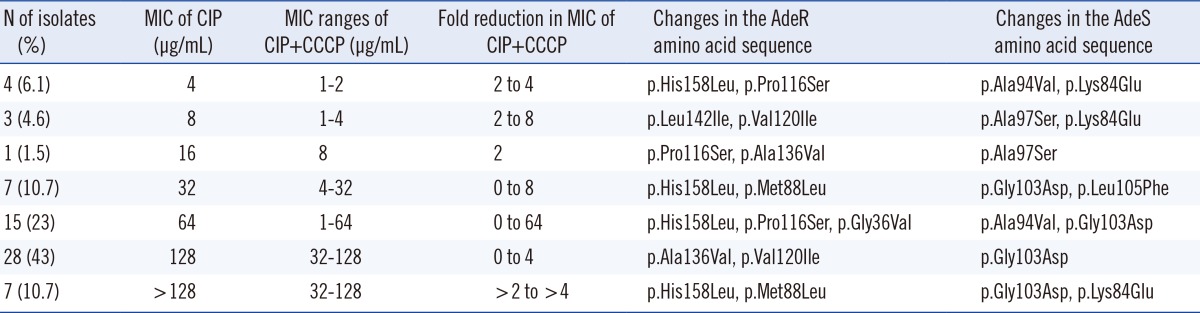
During the 6-month period, 68 isolates of A. baumannii were collected from hospitalized burn patients. Susceptibility testing using the disk diffusion method determined that 95.6% (65/68) of isolates showed resistance to CIP. According to the established breakpoint values recommended by the CLSI [20], A. baumannii isolates with MIC ≥4 µg/mL were considered CIP resistant. As shown in Table 1, the MIC of CIP in 6.1% (4/65) and 53.8% (35/65) of CRAB isolates was 4 µg/mL and ≥128 µg/mL, respectively, whereas a significant proportion of isolates (43%) had an MIC value equal to 128 µg/mL.
M-H agar plates containing 25 µg/mL CCCP were used to verify the efflux pump effect on CIP resistance in the 65 CRAB isolates that were found to possess adeRS-adeB genes. The addition of the EPI produced a 2- to 64-fold reduction of resistance in 86.2% (56/65) of the isolates that were resistant to CIP (Table 1). CCCP had no effect on the MIC of CIP for the A. baumannii ATCC 19606 strain. All bacteria grew well in M-H agar plates with CCCP but without CIP, indicating that 25 µg/mL CCCP had no intrinsic antibacterial activity. Interestingly, 3 CSAB isolates had no active efflux pump, as determined by the absence of changes in MIC after adding CCCP, despite the fact that they possessed relevant adeABC genes according to PCR results.
Specific PCR analysis demonstrated that all 65 CRAB and 3 CSAB isolates harbored the chromosomal adeB, adeR, and adeS genes, concomitantly. Amplification of transporter adeB, response regulator adeR, and its cognate kinase adeS genes produced bands of 541, 447, and 544 bp, respectively. To determine whether the CIP resistance in the 56 isolates harboring an active efflux pump was due to alterations in the regulatory region of the adeABC operon, we sequenced both amplified adeR and adeS genes. The nucleotide sequence data of the PCR productsshowed that 73.2% (41/56) of sequenced isolates had several point mutations in the AdeRS regulatory system. Point mutations in adeR (Pro116Leu) and adeS (Thr153Met) known to cause AdeABC overexpression were not identified in any isolates [13]. In addition, the isolates for which the MIC of CIP was higher (32 to ≥128 µg/mL) generally had more mutations in their adeRS genes. However, the sequencing results of PCR products demonstrated that the 3 CSAB isolates had no changes in adeR and adeS regulatory genes.
Prolonged patient stays in burn wards and the major threat of the spread of multidrug-resistant bacterial pathogens causing healthcare-associated infections are major concerns of infectious disease specialists [1, 22, 23]. A. baumannii is an important human pathogen that is able to survive on inanimate and dry surfaces for prolonged periods, and is isolated with increasing frequency from biological materials of burn patients. In recent years, this organism's resistance to various antibiotics, such as fluoroquinolones, has become a critical concern worldwide, including in Iran. In our study, 95.6% of A. baumannii isolates were resistant to CIP, similar to reports from other studies [24, 25, 26, 27]. However, the resistance rate observed in our investigation was higher than those found in the United Kingdom and China (50.9% and 61.2%, respectively) [6, 28]. This discrepancy could be due to differences in the quality control of antimicrobial susceptibility tests, patterns of antibiotic usage, environmental factors, and the geographical distribution of resistance in various countries. Most of the 65 resistant strains showed high levels of resistance to CIP, whereby the MIC range in 12.3% and 87.7% of these isolates was 4-16 µg/mL and 32 to more than 128 µg/mL, respectively. These results are consistent with the findings by Valentine et al. [5] where the MIC range of CIP of their isolates was 16-256 µg/mL [5]. Therefore, according to our results and those of other studies, the rate of resistance to CIP among A. baumannii strains has increased in many countries.
Various studies on A. baumannii and a number of other clinically important bacteria have demonstrated that efflux pumps play an important role in resistance to antibiotics, including fluoroquinolones [7, 29]. The 65 CRAB isolates evaluated in our study couldbe classified into two main groups: 1) positive for all three adeRS-adeB genes, with a conversed CIP resistance pattern in the presence of CCCP (86.2% of CRAB isolates), and 2) positive for all three adeRS-adeB genes, but no changes in MIC observed when an EPI was added to each M-H agar plate (13.8% of CRAB isolates). Consistent with the results by Lin et al. [30], we found that the MIC of CIP decreased in most CRAB isolates in the presence of CCCP, mainly 2- to 4-folds. As a four-fold or greater difference in susceptibility was considered significant, our study also showed that the MIC for 30 of the 65 resistant isolates (46.1%) was reduced significantly by 4- to 64-folds when the EPI was added. These results verified that enhanced efflux pumps are involved in resistance to fluoroquinolones in A. baumannii isolates. However, the failure of CCCP to affect the MICs of CRAB isolates in the second group, despite the presence of the AdeABC system, suggests that the efflux pump is not an important contributor to overt CIP resistance in some of our isolates. Instead, mutations in the GyrA and ParC subunits or other molecular mechanisms might be involved.
According to the nucleotide sequence data of the adeR and adeS genes, the first group of our CRAB isolates could be subdivided into two groups: a significant percentage (73.2%) of sequenced isolates that had several point mutations leading to amino acid substitutions in the AdeR and AdeS sequences, and a small number (26.8%) of isolates with no changes. Many of these mutations, including Leu142Ile and Val120Ile in adeR and Ala94Val and Phe214Leu in adeS, have been previously reported in multi-drug resistant isolates of A. baumannii [15, 31, 32]. In regard to their importance to drug resistance, the Ala94Val substitution is located within the histidine kinase, adenylyl cyclase, methyl-accepting chemotaxis protein and phosphatase (HAMP) domain of AdeS. The HAMP domain is thought to be involved in transmembrane signal transduction, and mutations in this domain have been associated with constitutive phenotypes [33, 34]. Therefore, both the conversion of the CIP resistance pattern in the presence of an EPI in all mutated isolates and the presence of changes within the adeRS operon of isolates that showed higher resistance suggest the possibility that multiple point mutations contribute to AdeABC pump overexpression and CIP-resistant phenotypes.
Although the presence of the AdeABC efflux pump was shown by PCR and verified by decreased CIP resistance in a phenotypic assay, we found no adeRS mutations in 26.8% of the sequenced isolates, a result that is consistent with the study by Peleg et al. [15]. The absence of mutations in these isolates, despite decreased MICs in the presence of CCCP, indicates either that other mechanisms (e.g., the insertion element ISAba1) might be responsible for increased pump activity or that other efflux systems are involved in CIP resistance.
In conclusion, our study represents the first attempt to demonstrate an increased rate of CRAB isolates as nosocomial pathogens and distributed AdeABC pumps as a mechanism associated with CIP resistance in the majority of the A. baumannii strains found in a clinical setting. Ongoing efforts should be aimed at detecting these bacteria and their resistance mechanisms, controlling their related infections, and employing new antimicrobials with activities against such problematic organisms. Our study may be limited by the facts that our experiments used bacterial isolates collected from a single clinical center and the expression level of adeB was not measured. Further studies are required to elucidate the precise mechanism leading to the enhanced pump activity and to determine the effect of ISAba1 insertion on the regulation of adeABC expression and drug resistance.
Acknowledgments
The work was financially supported by Iran University of Medical Sciences with grant number 1067. We gratefully acknowledge for partially conducting this study in microbiology department of Golestan University of Medical Sciences.
References
1. Azimi L, Motevallian A, EbrahimianNamvar A, Asghari B, Lari AR. Nosocomial infections in burned patients in motahari hospital, tehran, Iran. Dermatol Res Pract. 2011; 2011:436952. doi. 10.1155/2011/436952. Epub 2011 Nov 14. PMID: 22203838.

2. Lari AR, Alaghehbandan R, Nikui R. Epidemiological study of 3341 burns patients during three years in Tehran, Iran. Burns. 2000; 26:49–53. PMID: 10630320.

3. Sharma BR. Infection in patients with severe burns: causes and prevention thereof. Infect Dis Clin North Am. 2007; 21:745–759. PMID: 17826621.

4. National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004; 32:470–485. PMID: 15573054.
5. Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008; 46:2499–2507. PMID: 18524965.
6. Spence RP, Towner KJ. Frequencies and mechanisms of resistance to moxifloxacin in nosocomial isolates of Acinetobacter baumannii. J Antimicrob Chemother. 2003; 52:687–690. PMID: 12951327.
7. Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother. 2004; 54:821–823. PMID: 15355942.
8. Vila J, Ruiz J, Goñi P, Jimenez de Anta T. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J Antimicrob Chemother. 1997; 39:757–762. PMID: 9222045.
9. Park S, Lee KM, Yoo YS, Yoo JS, Yoo JI, Kim HS, et al. Alteration of gyrA, gyrB, and parC and activity of efflux pump in fluoroquinolone-resistant Acinetobacter baumannii. Osong Public Health Res Perspect. 2011; 2:164–170. PMID: 24159468.
10. Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv Drug Deliv Rev. 2005; 57:1486–1513. PMID: 15939505.

11. Zechini B, Versace I. Inhibitors of multidrug resistant efflux systems in bacteria. Recent Pat Antiinfect Drug Discov. 2009; 4:37–50. PMID: 19149695.

12. Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001; 45:3375–3380. PMID: 11709311.
13. Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004; 48:3298–3304. PMID: 15328088.
14. Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol. 2009; 191:2414–2418. PMID: 19136598.
15. Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007; 51:2065–2069. PMID: 17420217.
16. Ruzin A, Keeney D, Bradford PA. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother. 2007; 59:1001–1004. PMID: 17363424.
17. Bouvet PJ, Grimont PA. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol. 1986; 36:228–240.
18. Kuo HY, Yang CM, Lin MF, Cheng WL, Tien N, Liou ML. Distribution of blaOXA-carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn Microbiol Infect Dis. 2010; 66:195–199. PMID: 19836186.
19. Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006; 44:2974–2976. PMID: 16891520.
20. Clinical and Laboratory Standards Institute. Twentieth Informational supplement, M100-S120. Performance standards for antimicrobial susceptibility testing. Wayne, PA: CLSI;2010.
21. Pumbwe L, Glass D, Wexler HM. Efflux pump overexpression in multiple-antibiotic-resistant mutants of Bacteroides fragilis. Antimicrob Agents Chemother. 2006; 50:3150–3153. PMID: 16940115.
22. Azimi L, Rastegar Lari A, Alaghehbandan R, Alinejad F, Mohammadpoor M, Rahbar M. KPC-producer gram negative bacteria among burned infants in Motahari hospital, Tehran: first report from Iran. Ann Burns Fire Disasters. 2012; 25:74–77. PMID: 23233824.
23. Oncul O, Yüksel F, Altunay H, Açikel C, Celiköz B, Cavuşlu S. The evaluation of nosocomial infection during 1-year-period in the burn unit of a training hospital in Istanbul, Turkey. Burns. 2002; 28:738–744. PMID: 12464471.

24. Rahbar M, Mehrgan H, Aliakbari NH. Prevalence of antibiotic-resistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian J Pathol Microbiol. 2010; 53:290–293. PMID: 20551535.
25. Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006; 50:4114–4123. PMID: 17000742.
26. Lin MF, Liou ML, Tu CC, Yeh HW, Lan CY. Molecular epidemiology of integron-associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Ann Lab Med. 2013; 33:242–247. PMID: 23826559.
27. Asadollahi P, Akbari M, Soroush S, Taherikalani M, Asadollahi K, Sayehmiri K, et al. Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns. 2012; 38:1198–1203. PMID: 22579564.
28. Shi WF, Jiang JP, Xu N, Huang ZM, Wang YY. Inhibitory effects of reserpine and carbonyl cyanide m-chloro-phenylhydrazone on fluoroquinolone resistance of Acinetobacter baumannii. Chin Med J. 2005; 118:340–343. PMID: 15740676.
29. Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006; 19:382–402. PMID: 16614254.

30. Lin L, Ling BD, Li XZ. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int J Antimicrob Agents. 2009; 33:27–32. PMID: 18790612.
31. Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, et al. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother. 2010; 65:1589–1593. PMID: 20554571.
32. Sun JR, Chan MC, Chang TY, Wang WY, Chiueh TS. Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob Agents Chemother. 2010; 54:4934–4938. PMID: 20696871.
33. Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003; 185:89–97. PMID: 12486044.
34. Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007; 20:79–114. PMID: 17223624.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download