1. Mårdh PA, Tchoudomirova K, Elshibly S, Hellberg D. Symptoms and signs in single and mixed genital infections. Int J Gynaecol Obstet. 1998; 63:145–152.

2. Stamm WE. Chlamydia trachomatis infections: progress and problems. J Infect Dis. 1999; 179(S2):S380–S383.
3. Tait IA, Duthie SJ, Taylor-Robinson D. Silent upper genital tract chlamydial infection and disease in women. Int J STD AIDS. 1997; 8:329–331.

4. Taylor BD, Haggerty CL. Management of Chlamydia trachomatis genital tract infection: screening and treatment challenges. Infect Drug Resist. 2011; 4:19–29.

5. Bachmann LH, Johnson RE, Cheng H, Markowitz L, Papp JR, Palella FJ Jr, et al. Nucleic acid amplification tests for diagnosis of Neisseria gonorrhoeae and Chlamydia trachomatis rectal infections. J Clin Microbiol. 2010; 48:1827–1832.
6. Ossewaarde JM, Rieffe M, Rozenberg-Arska M, Ossenkoppele PM, Nawrocki RP, van Loon AM. Development and clinical evaluation of a polymerase chain reaction test for detection of Chlamydia trachomatis. J Clin Microbiol. 1992; 30:2122–2128.

7. Ridgway GL, Mumtaz G, Robinson AJ, Franchini M, Carder C, Burczak J, et al. Comparison of the ligase chain reaction with cell culture for the diagnosis of Chlamydia trachomatis infection in women. J Clin Pathol. 1996; 49:116–119.

8. Schepetiuk S, Kok T, Martin L, Waddell R, Higgins G. Detection of Chlamydia trachomatis in urine samples by nucleic acid tests: comparison with culture and enzyme immunoassay of genital swab specimens. J Clin Microbiol. 1997; 35:3355–3357.

9. Shattock RM, Patrizio C, Simmonds P, Sutherland S. Detection of Chlamydia trachomatis in genital swabs: comparison of commercial and in house amplification methods with culture. Sex Transm Infect. 1998; 74:289–293.

10. Van Dyck E, Ieven M, Pattyn S, Van Damme L, Laga M. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J Clin Microbiol. 2001; 39:1751–1756.
11. Watson EJ, Templeton A, Russell I, Paavonen J, Mardh PA, Stary A, et al. The accuracy and efficacy of screening tests for Chlamydia trachomatis: a systematic review. J Med Microbiol. 2002; 51:1021–1031.

12. Le Roy C, Papaxanthos A, Liesenfeld O, Mehats V, Clerc M, Bébéar C, et al. Swabs (dry or collected in universal transport medium) and semen can be used for the detection of Chlamydia trachomatis using the cobas 4800 system. J Med Microbiol. 2013; 62:217–222.

13. Mahony JB, Luinstra KE, Sellors JW, Jang D, Chernesky MA. Confirmatory polymerase chain reaction testing for Chlamydia trachomatis in first-void urine from asymptomatic and symptomatic men. J Clin Microbiol. 1992; 30:2241–2245.

14. Mania-Pramanik J, Donde UM, Maitra A. Use of polymerase chain reaction (PCR) for detection of Chlamydia trachomatis infection in cervical swab samples. Indian J Dermatol Venereol Leprol. 2001; 67:246–250.
15. Xiong L, Kong F, Zhou H, Gilbert GL. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J Clin Microbiol. 2006; 44:1413–1418.
16. Bandea CI, Kubota K, Brown TM, Kilmarx PH, Bhullar V, Yanpaisarn S, et al. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex Transm Infect. 2001; 77:419–422.

17. Claas HC, Melchers WJ, de Bruijn IH, de Graaf M, van Dijk WC, Lindeman J, et al. Detection of Chlamydia trachomatis in clinical specimens by the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1990; 9:864–868.
18. Mahony JB, Luinstra KE, Sellors JW, Chernesky MA. Comparison of plasmid- and chromosome-based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J Clin Microbiol. 1993; 31:1753–1758.

19. Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986; 16:52–62.

20. Land S, Tabrizi S, Gust A, Johnson E, Garland S, Dax EM. External quality assessment program for Chlamydia trachomatis diagnostic testing by nucleic acid amplification assays. J Clin Microbiol. 2002; 40:2893–2896.
21. Sevestre H, Mention J, Lefebvre JF, Eb F, Hamdad F. Assessment of Chlamydia trachomatis infection by Cobas Amplicor PCR and in-house LightCycler assays using PreservCyt and 2-SP media in voluntary legal abortions. J Med Microbiol. 2009; 58:59–64.

22. Verkooyen RP, Noordhoek GT, Klapper PE, Reid J, Schirm J, Cleator GM, et al. Reliability of nucleic acid amplification methods for detection of Chlamydia trachomatis in urine: results of the first international collaborative quality control study among 96 laboratories. J Clin Microbiol. 2003; 41:3013–3016.
23. Notomi T, Ikeda Y, Okadome A, Nagayama A. The inhibitory effect of phosphate on the ligase chain reaction used for detecting Chlamydia trachomatis. J Clin Pathol. 1998; 51:306–308.

24. Betsou F, Beaumont K, Sueur JM, Orfila J. Construction and evaluation of internal control DNA for PCR amplification of Chlamydia trachomatis DNA from urine samples. J Clin Microbiol. 2003; 41:1274–1276.
25. Chalker VJ, Vaughan H, Patel P, Rossouw A, Seyedzadeh H, Gerrard K, et al. External quality assessment for detection of Chlamydia trachomatis. J Clin Microbiol. 2005; 43:1341–1347.
26. Unemo M, Rossouw A, James V, Jenkins C. Can the Swedish new variant of Chlamydia trachomatis (nvCT) be detected by UK NEQAS participants from seventeen European countries and five additional countries/regions in 2009? Euro Surveill. 2009; 14(19):pii: 19206.

27. Lin JS, Jones WE, Yan L, Wirthwein KA, Flaherty EE, Haivanis RM, et al. Underdiagnosis of Chlamydia trachomatis infection. Diagnostic limitations in patients with low-level infection. Sex Transm Dis. 1992; 19:259–265.
28. Goessens WH, Mouton JW, van der Meijden WI, Deelen S, van Rijsoort-Vos TH, Lemmens-den Toom N, et al. Comparison of three commercially available amplification assays, AMP CT, LCx, and COBAS AMPLICOR, for detection of Chlamydia trachomatis in first-void urine. J Clin Microbiol. 1997; 35:2628–2633.

29. Helinski DR. Plasmids as vectors for gene cloning. Basic Life Sci. 1977; 9:19–49.

30. Meng S, Zhan S, Li J. Nuclease-resistant double-stranded DNA controls or standards for hepatitis B virus nucleic acid amplification assays. Virol J. 2009; 6:226.

31. Moncada J, Schachter J, Hook EW 3rd, Ferrero D, Gaydos C, Quinn TC, et al. The effect of urine testing in evaluations of the sensitivity of the Gen-Probe Aptima Combo 2 assay on endocervical swabs for Chlamydia trachomatis and neisseria gonorrhoeae: the infected patient standard reduces sensitivity of single site evaluation. Sex Transm Dis. 2004; 31:273–277.
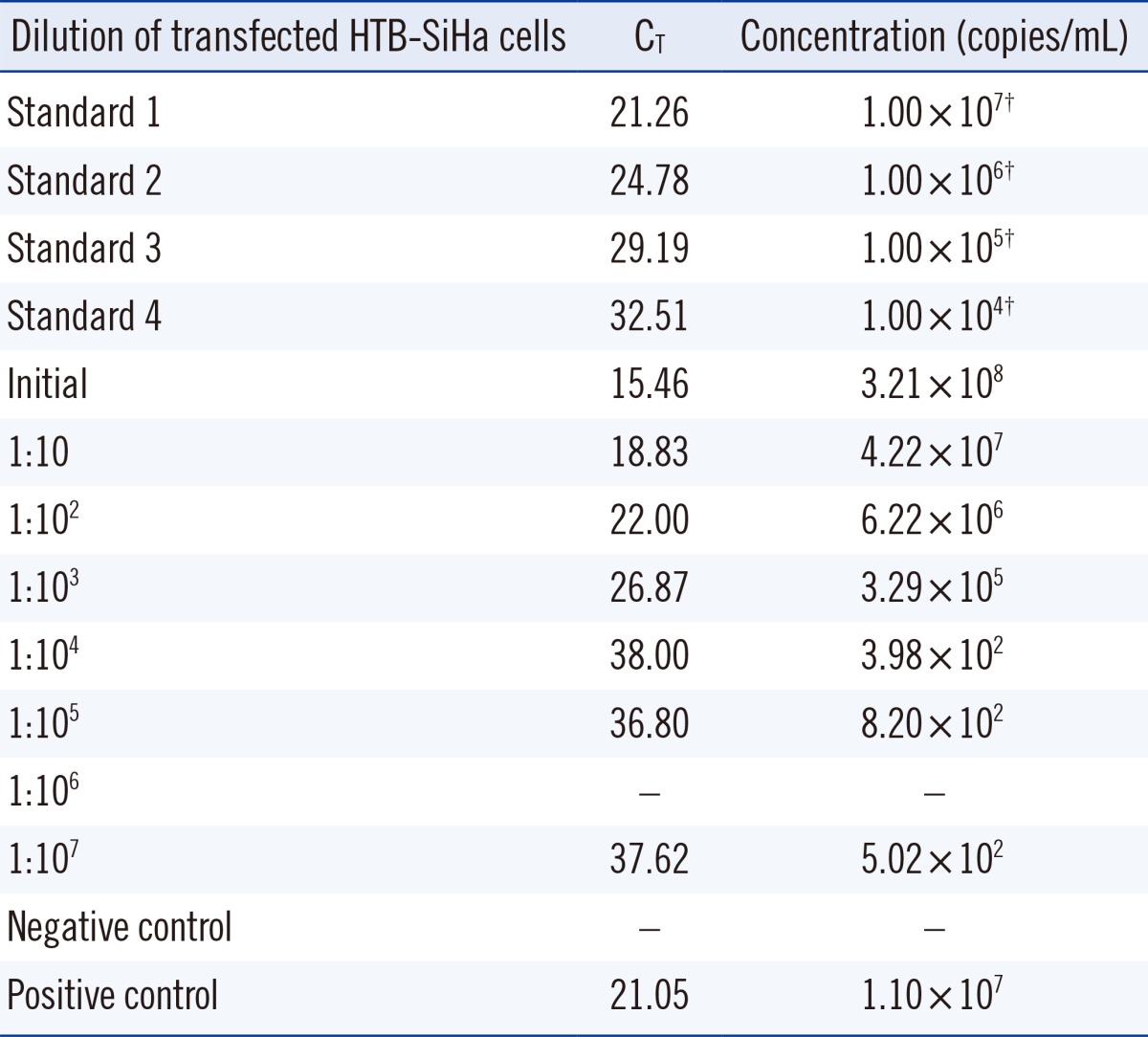
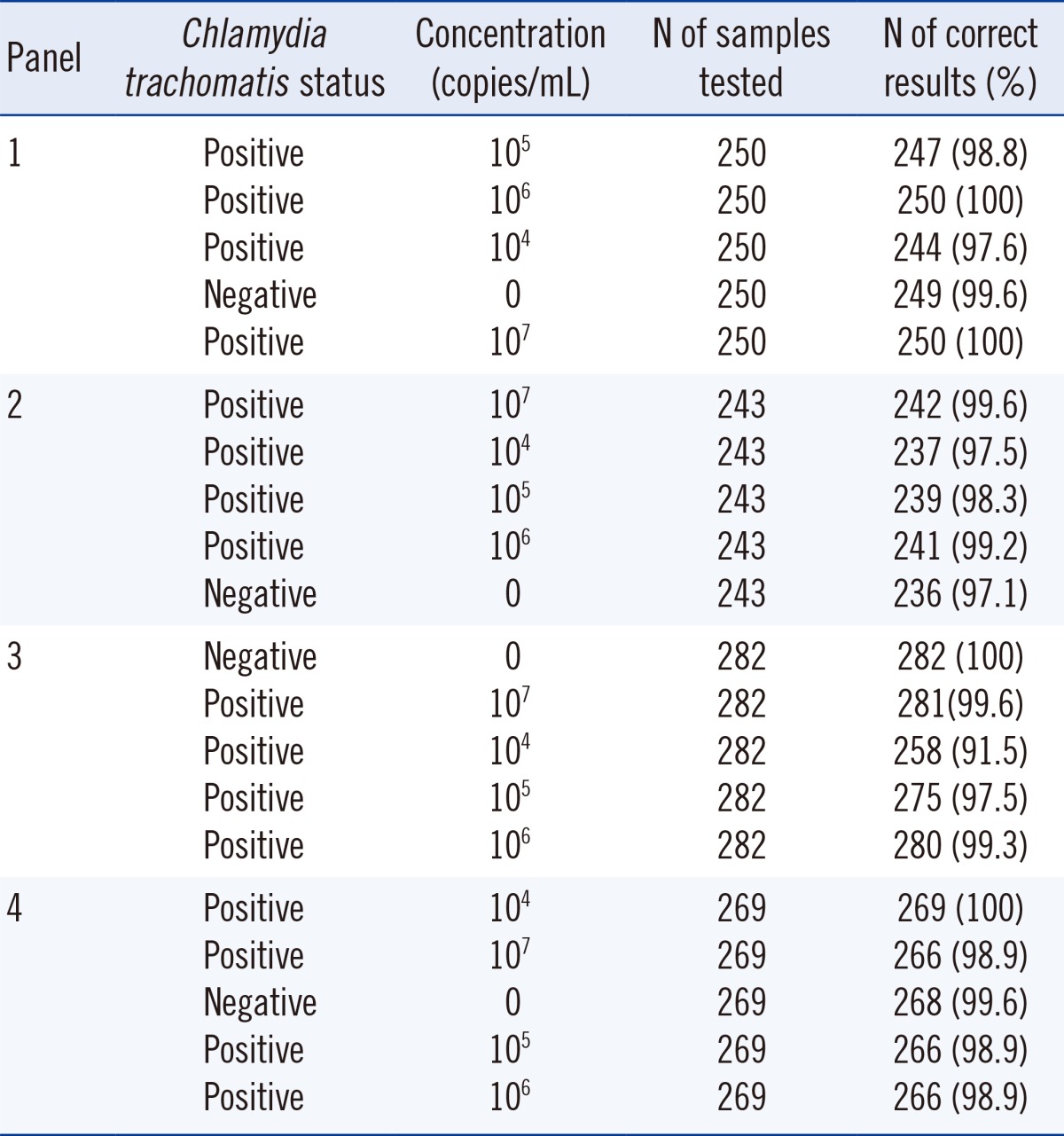
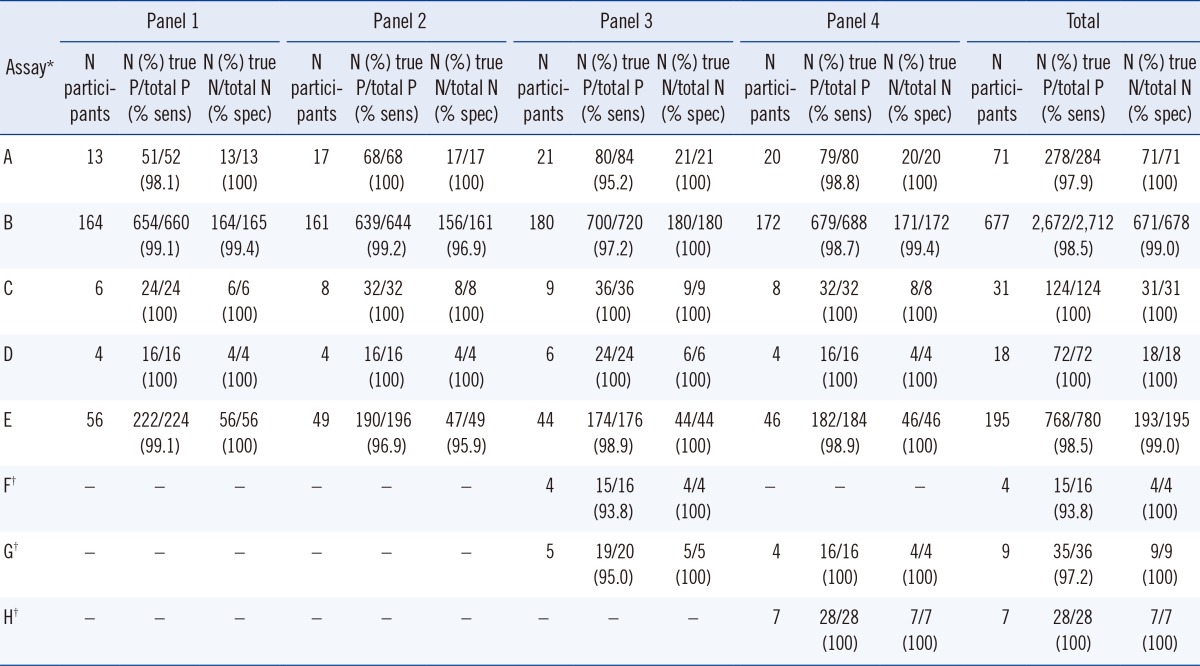




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download