Abstract
Background
Bacteria of the Mycobacterium abscessus group are the second most common pathogens responsible for lung disease caused by nontuberculous mycobacteria in Korea. There is still a lack of studies investigating the genetic mechanisms involved in M. abscessus resistance to antibiotics other than clarithromycin. This study investigated the characteristics of drug resistance exhibited by M. abscessus clinical isolates from Korea.
Methods
We performed drug susceptibility testing for a total of 404 M. abscessus clinical strains. Subspecies were differentiated by molecular biological methods and examined for mutations in drug resistance-related genes.
Results
Of the 404 strains examined, 202 (50.00%), 199 (49.26%), and 3 (0.74%) strains were identified as M. abscessus, M. massiliense, and M. bolletii, respectively. Of the 152 clarithromycin-resistant strains, 6 possessed rrl mutations, while 4 of the 30 amikacin-resistant strains contained rrs mutations, and 5 of the 114 quinolone-resistant strains had gyr mutations. All mutant strains had high minimal inhibitory concentration values for the antibiotics.
Mycobacterium abscessus-induced pulmonary disease accounts for approximately 65%-80% of the pulmonary diseases caused by rapidly growing mycobacteria [1-3]. On account of the resistance of M. abscessus against various antibiotics, pulmonary diseases are very difficult to treat [4-6]. A combination treatment (determined by M. abscessus
in vitro drug susceptibility testing) using specific antibiotics, such as amikacin, cefoxitin, imipenem, and macrolides, has been recommended by the American Thoracic Society and Infectious Disease Society of America [7]. However, the appropriate treatment duration has not yet been clearly established, and the cure rate is currently low. In Korea, M. abscessus pulmonary disease is the second most common pulmonary disease induced by nontuberculous mycobacteria [8, 9].
Recently, it was found that the M. abscessus group consists of M. abscessus (group I), M. massiliense, and M. bolletii (group II) strains [10]. A study also confirmed inducible resistance to clarithromycin in clinical strains of M. abscessus, in which the susceptibility to clarithromycin changed to resistance during in vitro drug susceptibility testing as the culture period progressed [11].
Furthermore, researchers have observed that the erythromycin ribosome methyltransferase (erm) gene is involved in the generation of inducible resistance to clarithromycin and that gene sequence variations between M. abscessus and M. massiliense strains are useful for bacterial identification in such cases [11, 12]. Additionally, inducible resistance has not been observed in M. massiliense, and the treatment outcome of M. massiliense infections with clarithromycin is better than that of M. abscessus [13]. On the basis of the inducible resistance of M. abscessus, the CLSI recently released its recommendations for analyzing the susceptibility test results of clarithromycin after a maximum incubation period of 14 days [14].
In contrast to earlier studies that have been performed with a limited number of strains, we investigated the distribution of M. massiliense and M. abscessus among the M. abscessus group clinical strains that the Korean Institute of Tuberculosis had received for nontuberculous mycobacteria identification and drug susceptibility testing and performed differential identification of the strains.
Because the mechanism of inducible resistance to clarithromycin in M. abscessus plays a role in the clarithromycin-based clinical outcomes, we evaluated the distribution and drug resistance characteristics of the strains that had acquired inducible resistance.
There is still an obvious lack of studies investigating the M. abscessus group genes involved in resistance to antibiotics other than clarithromycin. Therefore, this study aimed to contribute to the diagnosis and treatment of M. abscessus group infections by analyzing drug resistance against other antibiotics.
This study was conducted using 413 M. abscessus group clinical strains that were submitted for nontuberculous mycobacteria susceptibility testing from July 2009 to December 2010 at the Korean Institute of Tuberculosis. The selected strains were cultured in Lowenstein-Jensen medium. Of the 413 strains, 3 mixed strains and 6 contaminated strains were excluded from the study, and a total of 404 M. abscessus group clinical strains were analyzed.
Drug susceptibility was tested in 404 M. abscessus group clinical strains (202 M. abscessus strains, 199 M. massiliense strains, and 3 M. bolletii strains) using the micro-dilution method [14].
Approximately 100 µL/well of cation-adjusted Muller Hinton II (Becton Dickinson, San Jose, CA, USA) broth containing 50 mg/L 2,3-diphenyl-5-(2-thienyl)-tetrazolium chloride (STC: Tokyo Kasei Kogyo Co., Ltd., Tokyo, Japan) and 0.25 mg/L of clarithromycin, obtained by serial 0.5-fold dilutions from an initial concentration of 512 mg/L, was loaded onto microplate wells. The strains were then inoculated at a density of 104-105 cell/well, incubated at 30℃, and the minimal inhibitory concentration (MIC) was examined.
The strains were cultured for 3, 7, or 14 days and then the strains with MIC≤2 mg/L, MIC=4 mg/L, and MIC≥8 mg/L were considered susceptible, intermediate, and resistant, respectively. The strains showing clarithromycin susceptibility on day 3 and resistance after 7 days were determined to have inducible resistance.
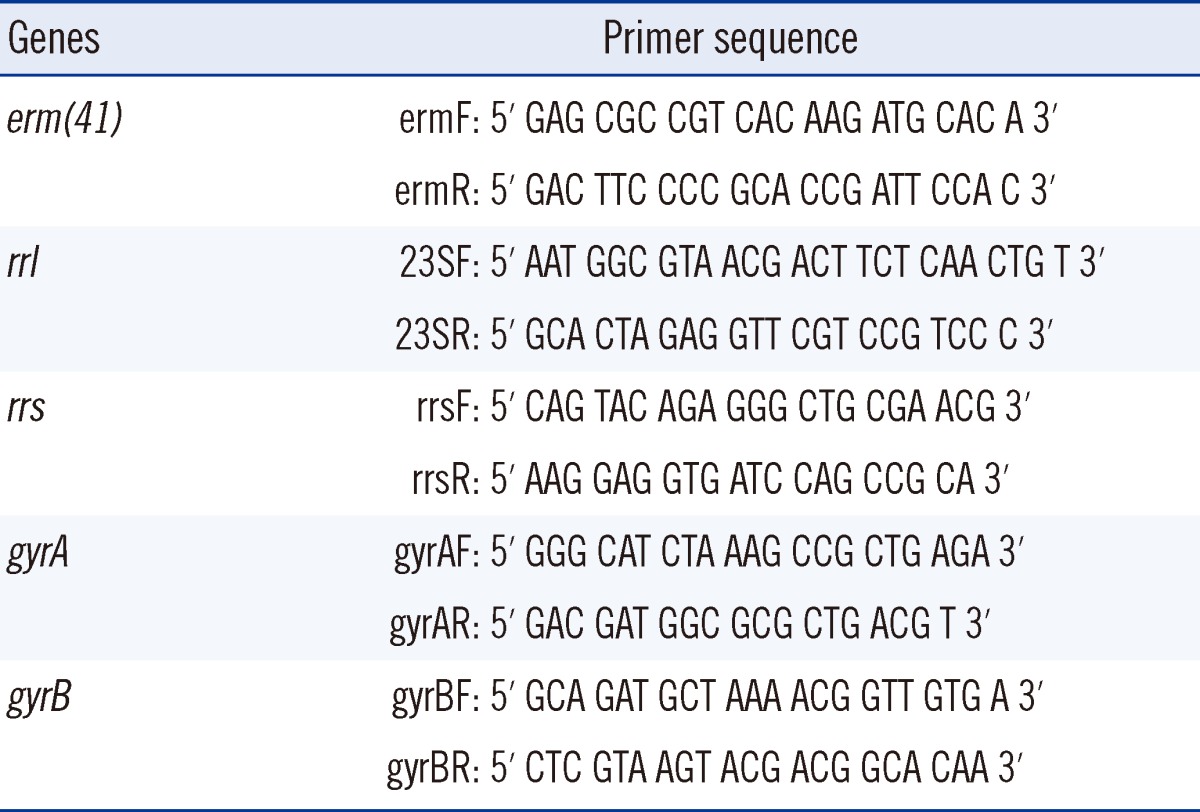
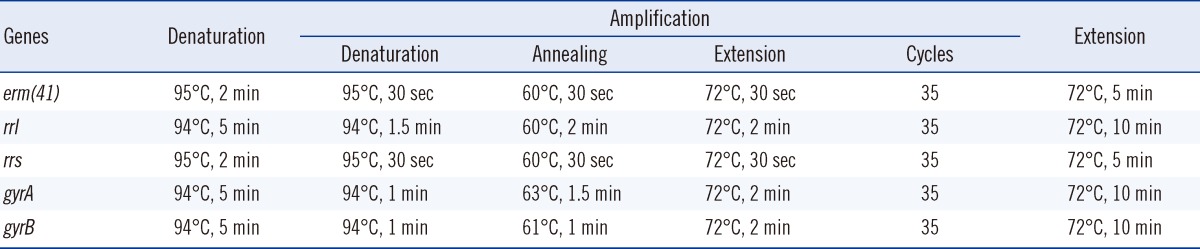
Acquired resistance to clarithromycin is associated with point mutations (at positions A2,058 and A2,059) in a region of the rrl gene encoding the peptidyltransferase domain of the 23S rRNA [15]. It has also been reported that the erm gene is associated with inducible resistance to clarithromycin. Therefore, characteristics of the susceptible and resistant clinical strains were compared and analyzed by sequencing the rrl and erm(41) genes. The primer sets and PCR conditions for amplification of two genes are outlined in Tables 1 and 2 [11, 16].
Drug susceptibility for amikacin was tested to investigate the distribution of the resistant strains. The strains were cultured for 3 days and strains with MIC≤16 mg/L, MIC=32 mg/L, and MIC≥64 mg/L were considered susceptible, intermediate, and resistant, respectively. The resistant strains were subjected to PCR and sequence analysis to confirm the base substitution (A→G) at position 1,408 (E. coli numbering) of the 16S ribosomal RNA (rRNA) gene rrs, which is an amikacin resistance-related gene (Tables 1 and 2) [17].
Drug susceptibility for ciprofloxacin and moxifloxacin was tested to examine the distribution of resistant strains. The strains were cultured for 3 days, and then strains with MIC≤1 mg/L, MIC=2 mg/L, and MIC≥4 mg/L were considered susceptible, intermediate, and resistant, respectively. We selected approximately one-third of the resistant strains with the highest MICs. PCR and sequence analysis were performed to investigate the mutations in the quinolone resistant-dependent region (QRDR) of the gyraseA (gyrA) and gyraseB (gyrB) genes (Tables 1 and 2) [18].
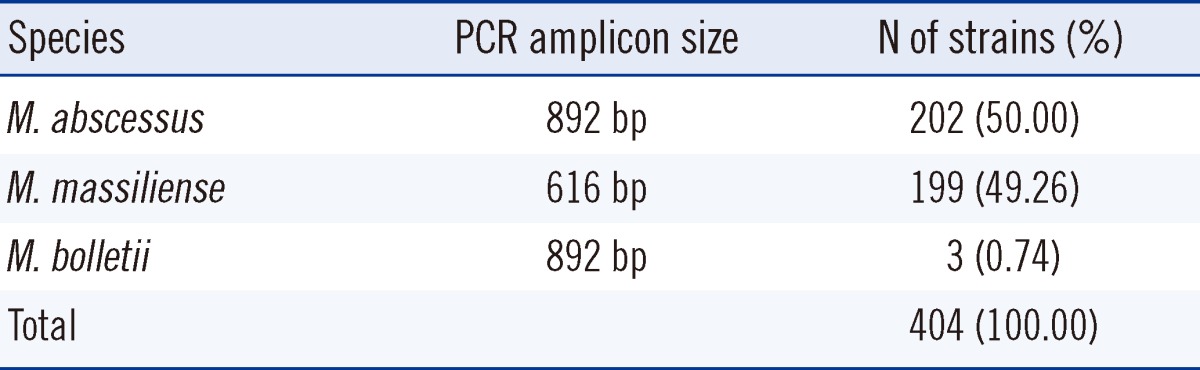
Amplification of the erm(41) gene resulted in an 892-bp PCR product for M. abscessus and a 616-bp product for M. massiliense, which is the erm(41)-deletion mutant. It was therefore possible to differentiate the 2 species according to their PCR product sizes. Additionally, M. bolletii was isolated from the M. abscessus strain and was separated by a -35 sequence difference in erm(41) gene promoter. Table 3 shows the distribution of the M. abscessus, M. massiliense, and M. bolletii clinical strains.
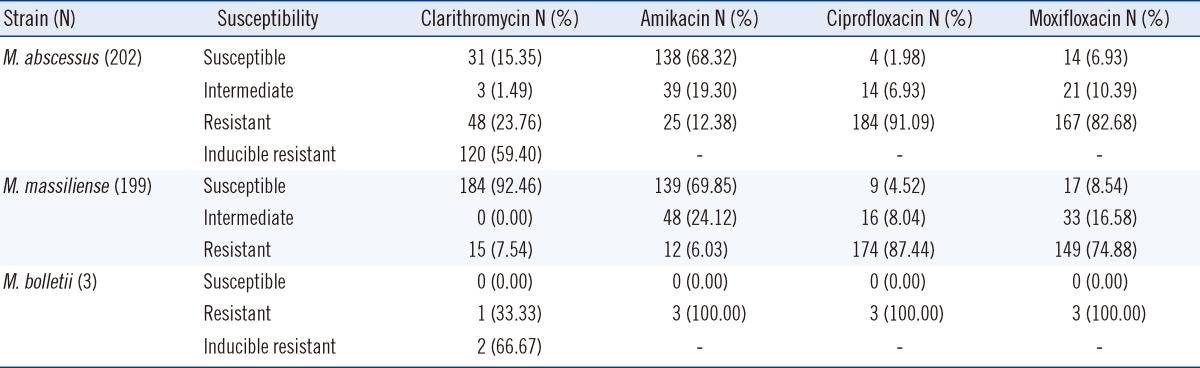
Clarithromycin susceptibility testing of 202 M. abscessus strains identified 31 susceptible stains, 48 resistant strains, 120 inducible resistance-bearing strains, and 3 intermediate strains. Indeed, most of the clinical strains (168 strains, 83%) were resistant to clarithromycin, which was most commonly utilized for treatment (Table 4). Among the 199 M. massiliense strains, 184 were susceptible and 15 were resistant, while among the 3 M. bolletii strains, 1 was resistant and 2 were inducible-resistant.
Of the 404 Mycobacterium abscessus group clinical strains, the sequencing results (for the rrl and erm(41) genes) were inconclusive for 51 strains, which were excluded from further analysis. Therefore, resistance-related gene analysis was performed on 157 M. abscessus strains, 194 M. massiliense strains, and 2 M. bolletii strains. Among the clarithromycin-resistant strains (140 M. abscessus, 12 M. massiliense, and 2 M. bolletii), 2 M. abscessus strains and 4 M. massiliense strains harbored point mutations in the peptidyltransferase domain of the 23S rRNA gene. These 6 strains showed resistance, with high MICs (>64 mg/L) (Table 5).
Thymine (T)/cytosine (C) point mutations were detected at position 28 of the M. abscessus
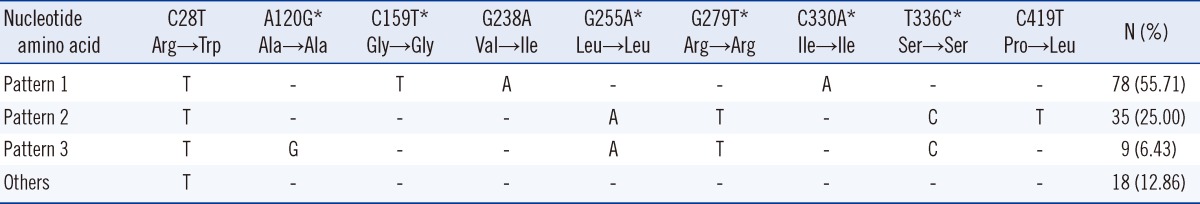
erm(41) gene. Of the 157 M. abscessus strains, 140 strains showed thymine 28 (T28 M. abscessus sequevar, Trp10 codon) mutations, while 17 had cytosine 28 (C28 M. abscessus sequevar, Arg10 codon) mutations. All the C28 strains were susceptible, whereas the T28 strains showed either resistance or inducible resistance. Of the 140 T28 M. abscessus strains, 78 strains presented an amino acid substitution at codon 80 (Val→Ile), and 35 strains presented an amino acid substitution at codon 140 (Pro→Leu; Table 6).
The amikacin resistant strains accounted for 69% (138 M. abscessus, 139 M. massiliense) of the study population (Table 4). Furthermore, rrs mutations were present in 2 M. abscessus and 2 M. massiliense strains. These 4 strains showed MIC values higher than 2,048 mg/L, thereby suggesting that the mutations occurred in highly resistant strains.
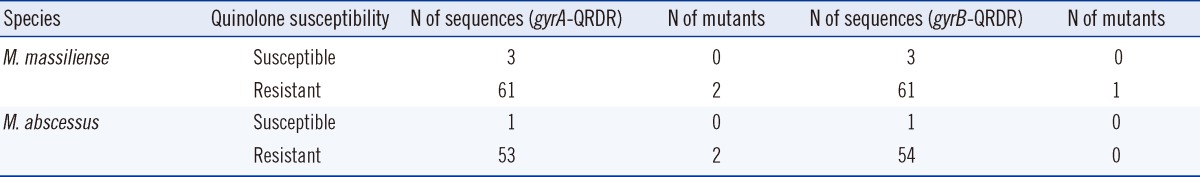
The strains resistant to ciprofloxacin or moxifloxacin accounted for over 70% (≥149 strains) of the study population (Table 4). In the mutation analysis of the QRDR in the gyrA and gyrB genes, 5 mutants were identified among the M. abscessus and M. massiliense resistant strains (Table 7). In terms of the gyrA gene, alanine at amino acid position 92 (M. abscessus numbering) was converted to valine in 1 strain, and aspartic acid at position 96 was mutated to asparagine in 3 strains. Arginine at amino acid position 492 of the gyrB gene was also converted to cysteine in 1 strain. Additionally, these 5 strains showed MIC values greater than 16 mg/L, thereby indicating that mutations are more likely to occur in the highly resistant strains.
Despite the continuous change in the taxonomic status of M. chelonae and M. abscessus, M. abscessus is considered as a separate species rather than a subspecies of M. chelonae. However, interspecific relationships have been identified within the M. abscessus group through genotype analysis, such as PCR restriction analysis (PRA) and sequencing of hsp65 and rpoB. Moreover, it has been recently reported that M. massiliense and M. bolletii are very closely related to M. abscessus [10-12, 19, 20]. In the present study M. abscessus and M. massiliense strains were differentiated on the basis of the erm(41) gene, and M. abscessus and M. bolletii were differentiated utilizing the -35 sequence difference in erm(41) gene promoter. As a result, out of the 202 strains of M. abscessus (50.00%), 199 strains of M. massiliense (49.26%) and 3 strains of M. bolletii (0.74%) were isolated, which is in good agreement with the distribution rate reported by Kim et al. [10].
Several mechanisms of antibiotic resistance have been proposed, including (1) changes in the target and receptors, (2) changes in membrane permeability, and (3) the active drug efflux pump [21-24].
In mycobacteria, clinically acquired macrolide resistance is caused by a point mutation at position 2,058 or 2,059 (E. coli numbering) in the 23S rRNA [25]. However, such mutations were rarely observed in the treatment of M. abscessus or M. chelonae infection [15]. According to a recent report, the erm(41) gene is involved in the acquisition of inducible-clarithromycin resistance by M. abscessus, thereby resulting in susceptibility at day 3 of incubation, and resistance after a maximum incubation period of 14 days. It was also confirmed that position 28 in the erm(41) gene in the resistant strains was mutated from C to T [11, 12, 26]. The results of the susceptibility testing of the 202 M. abscessus clinical strains demonstrated that the resistant strains accounted for 23.76% of the strains at day 3 of incubation, whereas their frequency increased to 59.40% after a maximum incubation period of 14 days. In addition, it was observed that among the 199 strains of M. massiliense, 184 strains were susceptible (92.5%) and 15 were resistant (7.5%); whereas among the M. bolletii, 1 was resistant, and 2 others showed inducible resistance. These results confirmed that the majority of the clinical strains of M. abscessus were resistant to clarithromycin (83.16%), which is a known therapeutic agent, and most of the clinical strains of M. massiliense were susceptible. These results support the findings of Koh et al. [13] that the treatment regimen containing clarithromycin was more effective in patients with M. massiliense pulmonary disease than in those with M. abscessus pulmonary disease, and the inducible resistance to clarithromycin shown in M. abscessus clinical strains played a role in the lack of efficacy of clarithromycin containing antibiotic therapy. Based on these results, the 2011 CLSI guidelines recommended that the incubation period of the strains be extended up to 14 days in the cases where the day 3 test indicates susceptibility [14].
In the analysis of the clarithromycin resistance-related 23S rRNA gene, 2 M. abscessus and 4 M. massiliense had rrl mutations, and both showed resistance at high MICs (MIC≥64 mg/L). Although it was not confirmed whether these strains had acquired drug resistance, the frequency of 23S rRNA mutants was low. In terms of the erm(41) sequence analysis, all resistant strains showed a T at nucleotide position 28, whereas all susceptible strains had a C (Table 6). Such results are in agreement with previous studies reporting that all erm(41) gene T28 type strains are resistant to clarithromycin [11, 12, 26]. Although the number of strains examined in this study was higher than that in previous studies, we could not identify any C28-type 23S rRNA mutants as reported by Bastian et al. [26]. It is noteworthy that 2 amino acid changes (Ile80 and Leu140 codon) were found in the erm(41) gene T28-type strains, except the Trp10 codon. Most of the resistant strains (80.71%) showed 2 distinct erm(41) gene sequence patterns that included silent mutations. Further studies need to be investigated whether these 2 amino acid changes facilitate resistance.
To the best of our knowledge, only a small number of studies have investigated the resistance characteristics of the drugs utilized for therapy, with the exception of clarithromycin. In this study, drug susceptibility and the characteristics of the drug resistance-related genes were examined to investigate the resistance to other drugs used for treatment.
In the amikacin susceptibility test, 6.03% of M. massiliense strains were resistant, whereas 12.38% of the strains in the M. abscessus showed resistance. Of the resistant strains, 4 mutations were found in both species when investigating the base mutation (A→G) at position 1,408 (E. coli numbering) of the 16S ribosomal RNA gene associated with amikacin resistance. In addition, the amikacin MIC values of all the mutant strains were remarkably high (>2,048 mg/L). Most of the strains showed susceptibility, and the distribution of the mutant strains was low. In previous reports, Prammananan et al. [17] identified that 16 out of the 17 examined M. abscessus strains were mutant with high MIC values because most strains were from the patients who had received aminoglycoside therapy. Another study also identified a new mutation that was not at the 1,408 position [27].
Fluoroquinolone antibiotics have been applied as effective therapeutic agents for infections induced by rapidly growing mycobacteria. Resistance to these antibiotics is mainly mediated by gyrA and gyrB gene mutations. Monego et al. [28] found that 31 out of 35 ciprofloxacin-resistant M. massiliense isolates showed mutations at amino acid position 90 (M. tuberculosis numbering, 92 M. abscessus numbering) but no mutation at position 94 (96 M. abscessus numbering) of gyrA. They stated that amino acid 90 of gyrA gene plays an important role in antibiotic resistance to fluoroquinolone. In this study, both M. massiliense and M. abscessus strains showed over 74% resistance to ciprofloxacin and moxifloxacin. Unlike previous studies, when investigating gyrA and gyrB mutations in one-third of the resistant strains, mutations at position 92 (alanine→valine, M. abscessus numbering), 96 of gyrA (aspartic acid→asparagine), and 492 of gyrB (arginine→cysteine) were observed in 1 strain (1 M. abscessus), 3 strains (1 M. abscessus, 2 M. massiliense), and 1 strain (1 M. massiliense), respectively. Amino acids at positions 90 and 94 in the A subunit (M. tuberculosis numbering system), and at positions 495, 516, and 533 in the B subunit (M. tuberculosis numbering) are frequently substituted in strains with acquired resistance to quinolones [18]. The mutation rate in this study was lower than that reported by Monego et al. [28], even though all the samples used in that study were collected for microbial culture before initial antibiotic treatment of patients and none of the patients has received quinolones for at least 4 weeks before the surgical procedures. They presumed that the incidence of ciprofloxacin-resistant M. massiliense may be due to selective pressure caused by drug abuse before the occurrence of the present cases. Further studies are required to fully establish the M. abscessus group susceptibility to fluoroquinolone antibiotics. Taken together, the results suggested that the mutations in the sequence encoding amino acid 96 of the gyrA gene and amino acid 492 of the gyrB gene are also involved in the resistance mechanisms, along with that encoding amino acid 92 of the gyrA gene (M. abscessus numbering).
In conclusion, we confirmed the characteristics of resistance related-genes in the M. abscessus group through drug susceptibility testing and analyses of resistance related-genes. The findings that most M. massiliense strains are susceptible to clarithromycin and amikacin, and most M. abscessus strains are susceptible to amikacin will aid the prescription of antibiotics for patients with infectious diseases.
References
1. Wallace RJ Jr, Swenson JM, Silcox VA, Good RC, Tschen JA, Stone MS. Spectrum of disease due to rapidly growing mycobacteria. Rev Infect Dis. 1983; 5:657–679. PMID: 6353528.

2. Griffith DE, Girard WM, Wallace RJ Jr. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis. 1993; 147:1271–1278. PMID: 8484642.

3. Han XY, Dé I, Jacobson KL. Rapidly growing mycobacteria: clinical and microbiologic studies of 115 cases. Am J Clin Pathol. 2007; 128:612–621. PMID: 17875513.
4. Jarand J, Levin A, Zhang L, Huitt G, Mitchell JD, Daley CL. Clinical and microbiologic outcomes in patients receiving treatment of Mycobacterium abscessus pulmonary disease. Clin Infect Dis. 2011; 52:565–571. PMID: 21292659.
5. Jeon K, Kwon OJ, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009; 180:896–902. PMID: 19661243.
6. Lyu J, Jang HJ, Song JW, Choi CM, Oh YM, Lee SD, et al. Outcomes in patients with Mycobacterium abscessuss pulmonary disease treated with long-term injectable drugs. Respir Med. 2011; 105:781–787. PMID: 21211956.
7. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial disease. Am J Respir Crit Care Med. 2007; 175:367–416. PMID: 17277290.
8. Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006; 129:341–348. PMID: 16478850.

9. Ryoo SW, Shin S, Shim MS, Park YS, Lew WJ, Park SN, et al. Spread of nontuberculous mycobacteria from 1993 to 2006 in Koreans. J Clin Lab Anal. 2008; 22:415–420. PMID: 19021271.

10. Kim HY, Kook Y, Yun YJ, Park CG, Lee NY, Shim TS, et al. Proportions of Mycobacterium massiliense and Mycobacterium bolletii strains among Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J Clin Microbiol. 2008; 46:3384–3390. PMID: 18753344.
11. Nash KA, Brown-Elliott BA, Wallace RJ Jr. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009; 53:1367–1376. PMID: 19171799.
12. Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, et al. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol Immunol. 2010; 54:347–353. PMID: 20536733.
13. Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011; 183:405–410. PMID: 20833823.
14. Clinical and Laboratory Standards Institute. M24-A2. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved Standard. Wayne, PA: CLSI;2011.
15. Wallace RJ Jr, Meier A, Brown BA, Zhang Y, Sander P, Onyi GO, et al. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996; 40:1676–1681. PMID: 8807061.
16. Nash KA, Inderlied CB. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995; 39:2625–2630. PMID: 8592991.
17. Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis. 1998; 177:1573–1581. PMID: 9607835.
18. Guillemin I, Jarlier V, Cambau E. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob Agents Chemother. 1998; 42:2084–2088. PMID: 9687411.

19. Adékambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006; 56:133–143. PMID: 16403878.
20. Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, et al. Amoebal coculture of "Mycobacterium massiliense" sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol. 2004; 42:5493–5501. PMID: 15583272.
21. Nash KA, Inderlied CB. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995; 39:2625–2630. PMID: 8592991.
22. Coleman K, Athalye M, Clancey A, Davison M, Payne DJ, Perry CR, et al. Bacterial resistance mechanisms as therapeutic targets. J Antimicrob Chemother. 1994; 33:1091–1116. PMID: 7928804.

23. Fierro JF, Hardisson C, Salas JA. Involvement if cell impermeability in resistance to macrolides in some producer streptomycetes. J Antibiot. 1988; 41:142–144. PMID: 3346187.
24. Banerjee SK, Bhatt K, Rana S, Misra P, Chakraborti PK. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem Biophys Res Commun. 1996; 226:362–368. PMID: 8806641.
25. Meier A, Heifets L, Wallace RJ Jr, Zhang Y, Brown BA, Sander P, et al. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J Infect Dis. 1996; 174:354–360. PMID: 8699066.
26. Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, et al. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother. 2011; 55:775–781. PMID: 21135185.
27. Nessar R, Reyrat JM, Murray A, Gicquel B. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J Antimicrob Chemother. 2011; 66:1719–1724. PMID: 21652621.
28. Monego F, Duarte RS, Biondo AW. gyrA and gyrB gene mutation in ciprofloxacin-resistant Mycobacterium massiliense clinical isolates from Southern Brazil. Microb Drug Resist. 2012; 18:1–6. PMID: 21711149.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download