Abstract
Background
Multidrug-resistant (MDR) Acinetobacter spp. acquire antimicrobial agent-resistance genes via class 1 integrons. In this study, integrons were characterized to investigate the antimicrobial resistance mechanisms of MDR Acinetobacter isolates. In addition, the relationship between the integron type and integron-harboring bacterial species was analyzed by using epidemiological typing methods.
Methods
Fifty-six MDR Acinetobacter spp.-A. baumannii (N=30), A. bereziniae (N=4), A. nosocomialis (N=5), and A. pittii (N=17)-were isolated. The minimum inhibitory concentrations (MICs) were determined on the basis of the results of the Epsilometer test (Etest). PCR and DNA sequencing was performed to characterize the gene cassette arrays of class 1 integrons. Multilocus sequence typing (MLST) and repetitive extragenic palindromic sequence (REP)-PCR were performed for epidemiological typing.
Results
Class 1 integrons were detected in 50 (89.3%) of the 56 isolates, but no class 2 or 3 integron was found within the cohorts. The class 1 integrons were classified into 4 types: 2.3-kb type A (aacA4-catB8-aadA1), 3.0-kb type B (aacA4-blaIMP-1-blaOXA-2), 3.0-kb type C (blaVIM-2-aacA7-aadA1), and 1.8-kb type D (aac3-1-blaOXA-2-orfD). Type A was most prevalent and was detected only in A. baumannii isolates, except for one A. bereziniae isolate; however, type B was amplified in all Acinetobacter isolates except for A. baumannii isolates, regardless of clone and separation time of the bacteria.
Acinetobacter spp. are aerobic, glucose-nonfermenting gram-negative bacteria that are widely distributed in soil and water environments [1, 2]. Isolates belonging to the A. calcoaceticus-A. baumannii (Acb) complex of the genus Acinetobacter are important nosocomial pathogens. Finding an appropriate antimicrobial therapy against these pathogens is a serious concern. The prevalence of Acb complex-related infections has markedly increased in recent years [3, 4, 5]. The Acb complex is composed of A. calcoaceticus, A. baumannii, A. pittii (formerly known as Acinetobacter genospecies), and A. nosocomialis (formerly known as Acinetobacter genospecies), which are genetically and phenotypically related. With the exception of A. calcoaceticus, all members of the Acb complex are commonly associated with hospital-acquired infections, and they rapidly acquire diverse antimicrobial resistance determinants [6, 7].
In Acinetobacter spp., the acquisition and dissemination of an antimicrobial-resistant determinant is frequently facilitated by integrons [8]. Integrons contain integrase gene (intI1), the product of which facilitates site-specific acquisitions and removals of the gene cassettes contained in the integrons. Approximately 9-17% of the sequenced bacterial genomes harbor integrons, and integrons with the same organization and composition have been found in unrelated isolates in spatially and temporally distinct areas [9, 10]. In particular, class 1 integrons, which are characterized by two conserved segments, are the most widely disseminated isolate of Acinetobacter spp. Class 1 integrons harbor various antimicrobial resistance gene cassettes encoding broad-spectrum β-lactamase, dihydroflavonol-4-reductase (dfr), and aminoglycoside-modifying enzymes (AMEs), such as acetyltransferase (aac), adenylyltransferase (aad), and phosphotransferase (aph) [11, 12, 13, 14, 15]. Therefore, multidrug-resistant (MDR) Acinetobacter spp. harboring genetic elements are resistant to three or more classes of antimicrobial agents that contain quinolones, aminoglycosides, ampicillin-sulbactams, extended-spectrum cephalosporins, and carbapenems [16].
Genetic elements like integrons frequently transfer antimicrobial resistance determinants, and are widely disseminated among MDR Acinetobacter spp.; however, scarce data is available about the epidemiological characterization of integron-harboring bacteria. In this study, integrons were characterized to investigate the antimicrobial resistance mechanisms of MDR Acinetobacter spp. identified in a university hospital in Daejeon, Korea over a period of 7 yr. In addition, multilocus sequence typing (MLST) and repetitive extragenic palindromic sequence (REP)-PCR were performed to analyze the relationship between the integron types and MDR Acinetobacter isolates harboring integrons.
A total of 56 consecutive and MDR Acinetobacter isolates were collected from a university hospital laboratory in Daejeon, Korea between January 2006 and December 2012. Acinetobacter spp. were identified by using the Vitek 2 Automated Instrument ID System (bioMérieux; Marcy l'Etoile, France) and by sequencing the partial rpoB housekeeping gene as described previously [17].
The MICs of Acinetobacter isolates for amikacin, gentamicin, ceftazidime, cefepime, imipenem, meropenem, and ciprofloxacin were determined by the Epsiolon test (Etest; bioMérieux). The data were interpreted as per the criteria approved by CLSI [18]. Escherichia coli ATCC 25922 was used as a reference strain.
Multiplex PCR was used to detect class 1, 2, and 3 integrons [19]. All of the integron-positive isolates were subjected to PCR and sequencing assays using specific primers for the analysis of gene cassette arrays. Class 1 integrons were amplified by using primers hep58 (5'-TCATGGCTTGTTATGACTGT-3') and hep59 (5'-GTAGGGCTTATTATGCACGC-3'). Primers hep74 (5'-CGGGATCCCGGACGGCATGCACGATTTGTA-3') and hep51 (5'-GATGCCATCGCAAGTACGAG-3') were used to amplify class 2 integrons [20]. Whole-cell (genomic) DNA was obtained from each target strain by using a genomic DNA purification kit (SolGent; Daejeon, Korea) according to the manufacturer's instructions. For sequencing, PCR products were purified with a PCR purification kit (SolGent) according to the manufacturer's instructions. Sequencing was performed by using the BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems; Foster City, CA, USA) and the ABI PRISM 3730XL DNA Analyzer (PE Applied Biosystems). DNA fragments (up to 1 kb in size) were sequenced by using the overlapping PCR technique. The various DNA sequences were confirmed on the basis of the BLAST paired alignment facility (http://blast.ncbi.nlm.gov).
For isolates of A. pittii spp., a MLST scheme with 7 housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) was used to determine the sequence types (STs) [21]. On the other hand, the Oxford MLST scheme with 7 housekeeping genes (cpn60, gdhB, gltA, gpi, gyrB, recA, and rpoD24) was used to determine the STs of A. baumannii isolates [22]. Each ST number was assigned by comparing the allele sequences to those in the MLST databases (http://www.pasteur.fr/mlst and http://pubmlst.org/abaumannii).
In addition, epidemiological typing of isolates was performed by REP-PCR [23]. In the REP-PCR method, the primer pair of REP1 (5'-III GCGCCGICATCAGGC-3') and REP2 (5'-ACGTCTTATCAGGCCTAC-3') was used to amplify putative REP-like elements in the genomic DNA. Amplification reactions were performed in a final volume of 50 µL, containing 100 ng of chromosomal DNA, 5 µL of 10× Taq buffer, 1.0 µL of 10 mM deoxyribonucleoside 5'-triphosphates (dNTPs) mix, 1.5 U of Taq DNA polymerase (SolGent), and 50 pmol of each primer. The cycling conditions were as follows: an initial denaturation step at 95℃ for 5 min followed by 30 cycles of 92℃ for 50 sec, 48℃ for 55 sec, and 70℃ for 5 min; and a final extension step at 70℃ for 10 min. The amplified products were separated via electrophoresis on 1.5% agarose gels containing ethidium bromide, and visualized by using the BioDoc-14TM Imaging system (UVP, Cambridge, UK).
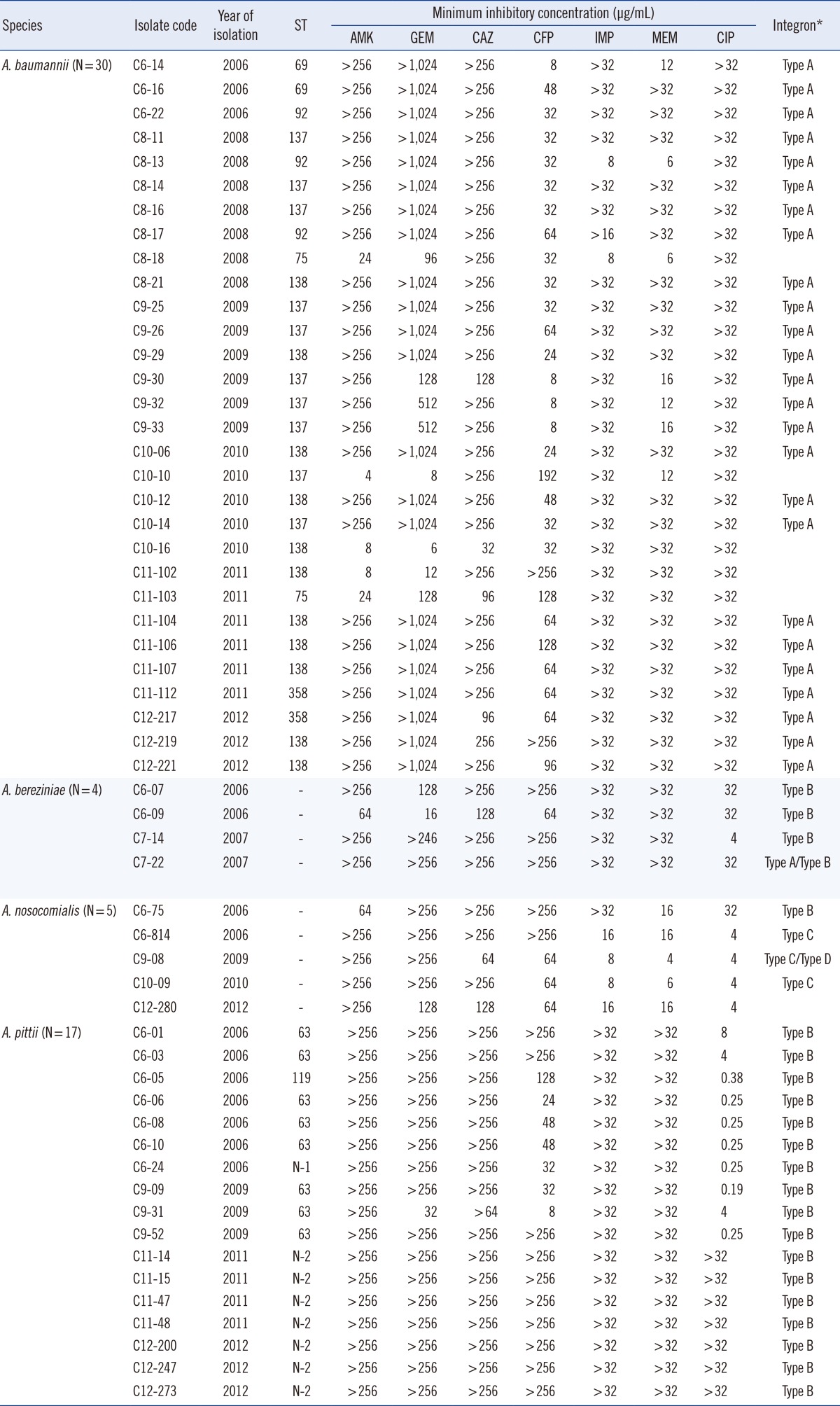
A total of 4 Acinetobacter spp. were identified: A. baumannii (N=30), A. bereziniae (N=4), A. nosocomialis (N=5), and A. pittii (N=17). The distribution of MICs for the 7 antimicrobial agents tested was similar among the 4 Acinetobacter isolates. Most of the Acinetobacter isolates showed high-level resistance to the antimicrobial agents, except for ciprofloxacin. MICs of ciprofloxacin were diverse, ranging from 0.19 mg/L to ≥32 mg/L (Table 1).
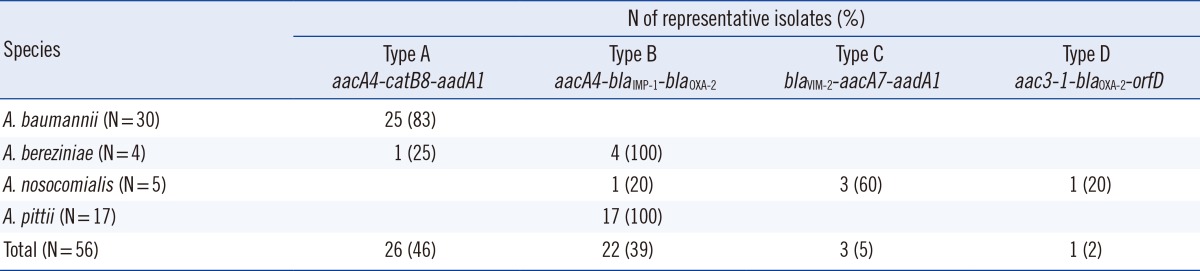
Class 1 integrons were detected in 50 (89.3%) of the 56 isolates, but no class 2 or 3 integron was found within the cohorts. The class 1 integrons were classified into 4 types (types A, B, C, and D) on the basis of the gene cassette nucleotide sequence (Table 1). The 2.3-kb type A (aacA4-catB8-aadA1) was the most prevalent type but was detected only in A. baumannii isolates, except for one in an A. bereziniae isolate (Table 2). The 3.0-kb type B (aacA4-blaIMP-1-blaOXA-2) was detected in 3 Acinetobacter spp. (A. bereziniae, A. nosocomialis, and A. pittii). The 3.0-kb type C (blaVIM-2-aacA7-aadA1) and 1.8-kb type D (aac3-1-blaOXA-2-orfD) were detected only in A. nosocomialis.
The A. baumannii isolates were grouped by MLST into 6 distinct STs, including ST69 (1-46-3-2-2-58-3), ST75 (1-3-3-2-2-11-3), ST92 (1-3-3-2-2-7-3), ST137 (1-3-3-2-2-12-3), ST138 (1-3-3-2-2-50-3), and ST358 (1-3-3-2-2-145-3). ST138 (n=11) was the most prevalent ST, followed by ST137 (n=10). In this study, the A. pittii isolates were grouped by MLST into 4 distinct STs, including ST63 (17-20-23-10-20-13-20), ST119 (36-20-38-16-38-18-20), and the novel STs N-1 (44-20-46-10-20-18-20) and N-2 (44-21-46-10-20-18-20). Two of the 4 STs corresponded to single isolates, whereas ST63 and N-2 corresponded to 8 and 7 strains, respectively.
All A. baumannii and A. pittii isolates were typed by REP-PCR. Although most A. baumannii isolates displayed the same REP-PCR type regardless of the STs, A. pittii isolates showed diverse patterns depending on the STs (Fig. 1).
In recent years, dissemination of the antimicrobial resistance genes through integrons in Acinetobacter spp. has become a major focus in the treatment of nosocomial infections by MDR bacteria [13]. Class 1 integrons have been found to be prevalent in Acinetobacter clinical isolates worldwide, which contain diverse gene cassettes that encode resistance factors to antimicrobial agents such as aminoglycosides, broad-spectrum β-lactams, and trimethoprims [14]. Therefore, our study was designed to determine the prevalence of various class 1 integrons of MDR Acinetobacter spp. In addition, we analyzed the relationship between the class 1 integron gene cassette array and Acinetobacter spp.
Most of the MDR Acinetobacter isolates (89.3%) analyzed in this study harbored class 1 integrons, and the frequency of these isolates in the study region was considerably higher than that in other geographical regions, including the United Kingdom (60%) and China (74.9%) [24]. However, the prevalence of class 1 integrons among MDR Acinetobacter spp. is similar to that documented previously showing the occurrence of class 1 integrase gene in only 15 of 123 Acinetobacter isolates (12%), with all of these 15 isolates showing MDR [25]. The higher detection rate of class 1 integrons was probably due to the pre-selection of clinical isolates showing the MDR phenotype. Our results indicate that class 1 integron is widely disseminated among MDR Acinetobacter spp. in the university hospital in Daejeon, Korea.
According to our results, most MDR Acinetobacter spp. carry class 1 integrons, but the specific integron type varies depending on the species. Type A is the most prevalent class 1 integron detected in Acinetobacter isolates. However, of the 4 Acinetobacter species tested, only an isolate belonging to A. baumannii harbored type A, except for one A. bereziniae isolate. Twenty-five A. baumannii strains harboring type A were collected between 2007 and 2012, and all of the strains belonged to 6 STs (ST69, ST75, ST92, ST137, ST138, and ST358). Our results suggest that type A is a dominant type of integron and that it has been conserved for many years in A. baumannii and other Acinetobacter species. The type A gene cassette array has been reported in A. baumannii as well as in other bacteria, including Acinetobacter spp. (92.0%; 126/137), Burkholderia cepacia (22.2%; 2/9), Klebsiella pneumonia (4.8%; 4/83), and E. coli (0.6%; 1/164) [24, 26]. Although the type A detection rate was highest in Acinetobacter isolates, there is a relative paucity of data on the non-A. baumannii Acinetobacter isolates harboring type A. These earlier reports support our results that type A can usually be transmitted horizontally and vertically among A. baumannii isolates.
In contrast to type A, type B was amplified in A. bereziniae, A. nosocomialis, and A. pittii isolates, but not in A. baumannii. In particular, all A. bereziniae and A. pittii isolates contained type B, irrespective of the time sampled or the clone. This result was similar to that of previous studies that found type B in A. pittii, A. bereziniae, and A. nosocomialis strains isolated from Taiwan and South Korea [27, 28]. Our results suggest that type B can usually be transmitted across species among non-A. baumannii Acinetobacter isolates, and is conserved in the Acinetobacter isolates.
In this study, we performed MLST and REP-PCR for epidemiological typing of MDR A. baumannii and A. pittii isolates. The STs were not directly correlated with REP-PCR types in the A. baumannii isolates, but they were well correlated in A. pittii isolates. Previous studies have reported no correlation of STs with REP-PCR types, probably because of the REP-PCR methods used; however, the MLST results were correlated with the REP-PCR clusters [29, 30]. Further studies are required to confirm the relationship between REP-PCR and MLST types. On the other hand, we confirmed that A. pittii exhibited a diverse REP-PCR pattern, which was less clonal than that of A. baumannii showing the same REP-PCR pattern.
All class 1 integrons detected in our study contained aminoglycoside-resistance genes such as acetyltransferase (aac) and adenylyltransferase (aad), which have been typically found within integron gene cassette arrays [13]. All isolates harboring class 1 integrons showed a high-level resistance rate to aminoglycoside compounds such as amikacin and gentamicin. In addition, the class 1 integrons harbored β-lactamase genes (blaOXA-2, blaIMP-1, and blaVIM-2) and chloramphenicol-resistance gene (catB8). Consequently, the co-occurrence of several antimicrobial-resistance determinants via class 1 integrons can lead to the emergence of MDR Acinetobacter spp. In particular, type B and type C harboring metallo-β-lactamase (MBL) genes (blaIMP-1 and blaVIM-2) were detected in non-A. baumannii Acinetobacter isolates. These results suggest that a major mechanism of carbapenem resistance in non-A. baumannii Acinetobacter isolates involves class 1 integrons containing MBL genes. However, in this study, integrons containing carbapenemase genes were not detected in the A. baumannii isolates. This result suggests that acquisitions of class 1 integrons were not considered as major factors of carbapenem resistance in A. baumannii isolates. It has been reported that the carbapenem resistance of A. baumannii isolates is often associated with the presence of carbapenem-hydrolyzing class D β-lactamase (CHDL) genes located in chromosomes, plasmids, or transposons [31].
Despite the increasingly frequent discovery of MDR Acinetobacter isolates in Korea, not much information is available regarding the distributions of integrons of MDR Acinetobacter strains isolated in Daejeon, Korea. We confirmed that class 1 integrons harboring diverse gene cassettes are widely disseminated among MDR Acinetobacter isolates. Although class 1 integrons can be transferred horizontally between unrelated isolates belonging to different species, each type of the 4 gene cassette arrays evaluated in this study was found in isolates belonging to Acinetobacter spp., irrespective of the time sampled and STs. Our findings suggest that certain class 1 integrons tend to transfer horizontally and vertically among A. baumannii or non-A. baumannii Acinetobacter isolates. Accordingly, the present study emphasizes that continuous analyses of gene cassette arrays of class 1 integrons will provide useful information regarding antimicrobial resistance mechanisms specific to different Acinetobacter species.
References
1. Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008; 46:1254–1263. PMID: 18444865.
2. Navon-Venezia S, Ben-Ami R, Carmeli Y. Update on Pseudomonas aeruginosa and Acinetobacter baumannii infections in the healthcare setting. Curr Opin Infect Dis. 2005; 18:306–313. PMID: 15985826.
3. Cisneros JM, Reyes MJ, Pachón J, Becerril B, Caballero FJ, García-Garmendía JL, et al. Bacteremia due to Acinetobacter baumannii: epidemiology, clinical findings, and prognostic features. Clin Infect Dis. 1996; 22:1026–1032. PMID: 8783704.
4. Sung JY, Kwon KC, Cho HH, Koo SH. Antimicrobial resistance determinants in imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolated in Daejeon, Korea. Korean J Lab Med. 2011; 31:265–270. PMID: 22016680.
5. Esterly JS, Griffith M, Qi C, Malczynski M, Postelnick MJ, Scheetz MH. Impact of carbapenem resistance and receipt of active antimicrobial therapy on clinical outcomes of Acinetobacter baumannii bloodstream infections. Antimicrob Agents Chemother. 2011; 55:4844–4849. PMID: 21825287.
6. Ko WC, Lee NY, Su SC, Dijkshoorn L, Vaneechoutte M, Wang LR, et al. Oligonucleotide array-based identification of species in the Acinetobacter calcoaceticus-A. baumannii complex in isolates from blood cultures and antimicrobial susceptibility testing of the isolates. J Clin Microbiol. 2008; 46:2052–2059. PMID: 18385442.
7. van den Broek PJ, van der Reijden TJ, van Strijen E, Helmig-Schurter AV, Bernards AT, Dijkshoorn L. Endemic and epidemic acinetobacter species in a university hospital: an 8-year survey. J Clin Microbiol. 2009; 47:3593–3599. PMID: 19794057.
8. Gootz TD, Marra A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther. 2008; 6:309–325. PMID: 18588496.
9. Machado E, Coque TM, Cantón R, Sousa JC, Peixe L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J Antimicrob Chemother. 2008; 62:296–302. PMID: 18456652.

10. Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010; 44:141–166. PMID: 20707672.
11. Da Silva GJ, Correia M, Vital C, Ribeiro G, Sousa JC, Leitão R, et al. Molecular characterization of blaIMP-5, a new integron-borne metallo-β-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol Lett. 2002; 215:33–39. PMID: 12393197.
12. Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C, et al. Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob Agents Chemother. 2002; 46:3665–3668. PMID: 12384388.
13. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004; 53:1233–1240. PMID: 15585503.
14. Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006; 4:608–620. PMID: 16845431.

15. Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–190. PMID: 19361944.
16. Srinivasan VB, Rajamohan G, Pancholi P, Stevenson K, Tadesse D, Patchanee P, et al. Genetic relatedness and molecular characterization of multidrug resistant Acinetobacter baumannii isolated in central Ohio, USA. Ann Clin Microbiol Antimicrob. 2009; 8:21. PMID: 19531268.

17. Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009; 155:2333–2341. PMID: 19389786.
18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth Informational supplement, M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute;2010.
19. Dillon B, Thomas L, Mohmand G, Zelynski A, Iredell J. Multiplex PCR for screening of integrons in bacterial lysates. J Microbiol Methods. 2005; 62:221–232. PMID: 16009279.

20. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001; 45:2658–2661. PMID: 11502548.
21. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010; 5:e10034. PMID: 20383326.
22. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005; 43:4382–4390. PMID: 16145081.
23. Bou G, Cerveró G, Domínguez MA, Quereda C, Martínez-Beltrán J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000; 6:635–643. PMID: 11284921.
24. Wu K, Wang F, Sun J, Wang Q, Chen Q, Yu S, et al. Class 1 integron gene cassettes in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents. 2012; 40:264–267. PMID: 22817917.

25. Golanbar GD, Lam CK, Chu YM, Cueva C, Tan SW, Silva I, et al. Phenotypic and molecular characterization of Acinetobacter clinical isolates obtained from inmates of California correctional facilities. J Clin Microbiol. 2011; 49:2121–2131. PMID: 21450955.
26. Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, et al. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol. 2005; 43:3074–3082. PMID: 16000417.
27. Lin YC, Sheng WH, Chen YC, Chang SC, Hsia KC, Li SY. Differences in carbapenem resistance genes among Acinetobacter baumannii, Acinetobacter genospecies 3 and Acinetobacter genospecies 13TU in Taiwan. Int J Antimicrob Agents. 2010; 35:439–443. PMID: 20106635.
28. Park YK, Jung SI, Park KH, Kim SH, Ko KS. Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea. Int J Antimicrob Agents. 2012; 39:81–85. PMID: 21996405.
29. Higgins PG, Janssen K, Fresen MM, Wisplinghoff H, Seifert H. Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol. 2012; 50:3493–3500. PMID: 22895032.
30. Park S, Kim HS, Lee KM, Yoo JS, Yoo JI, Lee YS, et al. Molecular and epidemiological characterization of carbapenem-resistant Acinetobacter baumannii in non-tertiary Korean hospitals. Yonsei Med J. 2013; 54:177–182. PMID: 23225816.
31. Roca I, Espinal P, Vila-Farrés X, Vila J. The Acinetobacter baumannii Oxymoron: Commensal Hospital Dweller Turned Pan-Drug-Resistant Menace. Front Microbiol. 2012; 3:148. PMID: 22536199.

Fig. 1
Repetitive extragenic palindromic (REP)-PCR patterns of genomic DNA from MDR Acinetobacter baumannii (top panel) and Acinetobacter pittii (bottom panel) isolates. Lane M, 1-kb DNA size marker.
Abbreviations: MDR, multidrug resistant; ST, sequence type.
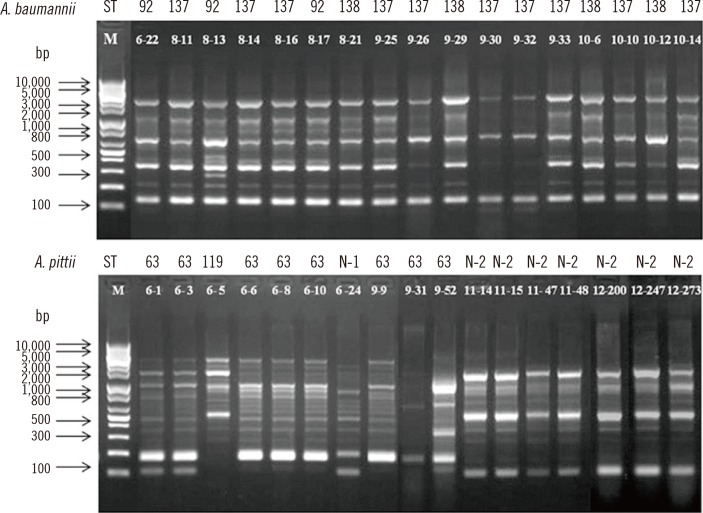
Table 1
The MIC distribution of 7 antimicrobial agents for 56 MDR Acinetobacter spp. isolates
*Type A, aacA4-catB8-aadA1; Type B, aacA4-blaIMP-1-blaOXA-2; Type C, blaVIM-2-aacA7-aadA1; Type D, aac3-1-blaOXA-2-orfD.
Abbreviations: MIC, minimum inhibitory concentration; ST, sequence type; AMK, amikacin; GEN, gentamicin; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; N, novelsequence type.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download