Abstract
We assessed the reporting times for identification of nasal methicillin-resistant Staphylococcus aureus (MRSA) carriers in 2011 in a university-affiliated hospital using surveillance cultures incubated for 1 and 2 days with ChromID MRSA (bioMérieux, France). Of 2,732 nasal swabs tested, MRSA was detected in 829 (85.6%) and 140 (14.4%) swabs after 1 and 2 days of incubation, respectively, and the median reporting times for positive specimens were 33.7 hr (range, 18.2-156.9 hr) and 108.1 hr (range, 69.8-181.0 hr), respectively. Detection rate after 1-day incubation was 85%. Additional 1-day incubation improved detection rate; however, it prolonged the reporting times of positive specimens approximately up to 4 days because of the need for confirmatory tests such as species identification and susceptibility tests. Following a 2-day culture with ChromID MRSA, rapid confirmatory tests are warranted to reduce delay in identifying MRSA carriers.
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of healthcare-associated infections worldwide. Healthcare-associated MRSA infections are associated with increased mortality, morbidity, and excess hospital costs [1, 2]. In Korea, MRSA accounts for generally 70% of clinical isolates of S. aureus since the mid-1990s [3]. Hence, active screening to detect asymptomatic carriers is recommended as a part of an effective measure to prevent nosocomial infections [4, 5]. The use of chromogenic agar is a simple and low-cost MRSA screening method and more rapid than using traditional cultures [6, 7]. Generally, an incubation time of 18-24 hr and 48 hr is recommended for the detection of MRSA with the currently available chromogenic media, according to the manufacturer's instructions [6, 8, 9]. However, the incubation period is longer than that, during weekends and holidays in clinical laboratories that perform batch tests for bacterial cultures. In addition, there is a lack of published data regarding the effect of longer incubation during weekends and holidays, as well as the optimal length of incubation in clinical settings. In this study, we investigated the time interval between specimen submission and laboratory reporting of MRSA and compared the reporting time for working days and calendar days (total days) by using ChromID MRSA (bioMérieux, Marcy l'Etoile, France).
Between January and December 2011, 2,732 specimens were collected from the anterior nares of patients in the surgical and medical intensive care units on admission at a 720-bed university hospital. The specimen-receiving area was open for 24 hr, but the samples were inoculated twice a day from Monday to Friday. During weekends and holidays, specimens were inoculated only once a day. Submitted specimens were directly inoculated on ChromID MRSA agar. Plates were incubated at 35℃ in ambient air and read on the next day at 8 a.m. after 18-24 hr of incubation. According to the manufacturer's instructions, green-colored colonies observed after 18-24 hr of incubation were presumed to be MRSA. We reported such cultures as "MRSA isolated-presumptive identification" followed by subculture on blood agar plates (BAP) for confirmation. On BAP, colonies morphologically consistent with S. aureus were tested with MicroScan Pos Breakpoint Combo Panel Type 28 (Siemens Healthcare Diagnostics Inc., West Sacramento, CA, USA) by using MicroScan Walk-Away 96 (Siemens Healthcare Diagnostics Inc.). Other colonies morphologically incompatible with S. aureus were tested with latex agglutination reagent (Pastorex Staph Plus test, Bio-Rad, Marne la Coquette, France), and positive results were confirmed by MicroScan. Plates interpreted as negative on the first day were reincubated for an additional day. As the manufacturer recommended that identification of characteristic colonies be initially confirmed using biochemical or immunological tests (S. aureus) and then, methicillin resistance be checked after 48 hr of incubation, we performed a biochemical identification test and a methicillin resistance test by using MicroScan after a 2-day incubation. In our laboratory, colony reading was not performed at weekends and holidays. Therefore, specimens inoculated on Sunday to Thursday were designated as being within the workday schedule, and the others were designated as being out of the workday schedule. We investigated the reporting time for specimens both within and out of the workday schedule.
In all, 974 (35.7%) green-colored colonies were detected from 2,732 specimens, five of which were determined to be non-MRSA by MicroScan or the latex agglutination test. Eight hundred and twenty-nine (85.6%) of the MRSA isolates were detected on the first reading day and 140 (14.4%) were detected on the second reading day. The median reporting time was 33.7 hr (range, 18.2-156.9 hr) and 108.1 hr (range, 69.8-181.0 hr), respectively (Fig. 1). As mentioned above, five samples that were positive on ChromID MRSA were revealed as non-MRSA isolates, including two isolates of Staphylococcus haemolyticus and one isolate of methicillin susceptible S. aureus (minimum inhibitory concentration [MIC] <0.25 µg/mL). The remaining two isolates were negative in the latex agglutination test, and further tests were not performed. The median reporting time for non-MRSA isolates was 33.8 hr (range, 24.7-41.5 hr).
There were 1,882 specimens examined within the workday schedule. Of these, 653 (34.7%) green colonies, including the five non-MRSA isolates, were detected. Among the 648 (34.4%) MRSA isolates, 571 (88.1%) samples were detected after 1 day of incubation. The median reporting time was 32.9 hr (range, 19-55.2 hr). An additional 77 (11.9%) MRSA isolates were detected after 2 days of incubation, and the median reporting time was 104.1 hr (range, 69.8-165.5 hr; Fig. 1). Some plates were examined after the recommended 48-hr incubation, because we did not perform the routine experiments during holidays and weekends owing to our laboratory working policies. This delay in interpretation is one of the limitations in this study. Modification of the laboratory policy to enable interpretation during holidays could minimize this problem.
In this study, 829 (85.6%) out of 969 MRSA isolates were recovered on the first reading day. Several studies of ChromID MRSA reported different sensitivities and specificities. In two studies, for cultures incubated for 16-18 hr, the sensitivity was 51% or less, and sensitivity increased to 82% and 75% after 42 hr of incubation [10, 11]. Similarly, another study reported an improved sensitivity from 48.6% to 71.3% after increasing the incubation time from 16-23 hr to 22-24 hr, and specificities after 24-hr incubation were equivalently high regardless of the incubation time [8]. In accordance with the previous studies, our results showed increased detection rate after 2 days of incubation. We tested more than 800 chromogenic positive colonies with MicroScan, and only five samples were identified as non-MRSA. This supports the manufacturer's recommendation that additional tests to confirm MRSA are not required after 18-24 hr of incubation.
Chromogenic media showed a wide range of reporting times in the hospital setting. On average, colonies detected after overnight incubation could be reported within 1 or 2 days. However, colonies detected after a 2-day incubation required an additional 47.2 hr for subculture and further biochemical tests, which resulted in a reporting time of around 2 days compared with overnight incubation. Overall, 140 (14.4%) MRSA isolates were additionally detected after a 2-day incubation and were reported as MRSA 4 or 5 days after the specimen had been received. Such a long reporting time results in a delay in identifying MRSA carriers and providing timely management. In the worst case in our study, reporting was done 7 days after the specimen had been received because of long or continuous holidays. The delay in reporting time of approximately 3 days was mainly caused by the need for additional biochemical tests for identification of S. aureus and its methicillin-resistance. To reduce the reporting time after a 2-day incubation, it might be useful to perform the latex agglutination test instead of the biochemical test for identification of S. aureus or to use chromogenic agar media with an incubation time of 24 hr. Recently, a new version of ChromID MRSA has been developed that can be read within 24 hr [12].
In conclusion, ChromID MRSA requires a 2-day incubation for optimal detection rate. MRSA nasal carriers, however, might be identified after 4 days, if biochemical confirmatory tests are used following a 2-day incubation on ChromID MRSA. Other approaches should be considered to reduce the reporting time for identification of MRSA carriers.
References
1. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003; 36:53–59. PMID: 12491202.
2. Engemann JJ, Carmeli Y, Cosgrove SE, Fowler VG, Bronstein MZ, Trivette SL, et al. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin Infect Dis. 2003; 36:592–598. PMID: 12594640.
3. Lee H, Kim CK, Lee J, Lee SH, Ahn JY, Hong SG, et al. Antimicrobial resistance of clinically important bacteria isolated from 12 hospitals in Korea in 2005 and 2006. Korean J Clin Microbiol. 2007; 10:59–69.
4. Tacconelli E, De Angelis G, de Waure C, Cataldo MA, La Torre G, Cauda R. Rapid screening tests for meticillin-resistant Staphylococcus aureus at hospital admission: systematic review and meta-analysis. Lancet Infect Dis. 2009; 9:546–554. PMID: 19695491.
5. Harbarth S, Hawkey PM, Tenover F, Stefani S, Pantosti A, Struelens MJ. Update on screening and clinical diagnosis of meticillin-resistant Staphylococcus aureus (MRSA). Int J Antimicrob Agents. 2011; 37:110–117. PMID: 21163628.
6. Malhotra-Kumar S, Haccuria K, Michiels M, Ieven M, Poyart C, Hryniewicz W, et al. Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococcus species. J Clin Microbiol. 2008; 46:1577–1587. PMID: 18322065.
7. Wassenberg MW, Kluytmans JA, Box AT, Bosboom RW, Buiting AG, van Elzakker EP, et al. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin Microbiol Infect. 2010; 16:1754–1761. PMID: 20219077.
8. Morris K, Wilson C, Wilcox MH. Evaluation of chromogenic meticillin-resistant Staphylococcus aureus media: sensitivity versus turnaround time. J Hosp Infect. 2012; 81:20–24. PMID: 22463978.
9. Carson J, Lui B, Rosmus L, Rennick H, Fuller J. Interpretation of MRSASelect screening agar at 24 hours of incubation. J Clin Microbiol. 2009; 47:566–568. PMID: 19144795.
10. Nahimana I, Francioli P, Blanc DS. Evaluation of three chromogenic media (MRSA-ID, MRSA-Select and CHROMagar MRSA) and ORSAB for surveillance cultures of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2006; 12:1168–1174. PMID: 17121622.
11. Nonhoff C, Denis O, Brenner A, Buidin P, Legros N, Thiroux C, et al. Comparison of three chromogenic media and enrichment broth media for the detection of methicillin-resistant Staphylococcus aureus from mucocutaneous screening specimens: comparison of MRSA chromogenic media. Eur J Clin Microbiol Infect Dis. 2009; 28:363–369. PMID: 18855028.
12. Van Hoecke F, Deloof N, Claeys G. Performance evaluation of a modified chromogenic medium, ChromID MRSA New, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. Eur J Clin Microbiol Infect Dis. 2011; 30:1595–1598. PMID: 21499707.
Fig. 1
Time interval between specimen submission and laboratory reporting time for methicillin-resistant Staphylococcus aureus nasal swabs. Data were sorted into total day (□) and working day ( ). The bottom and top edges of the boxes mark the first quartile and third quartile, respectively. The horizontal line in the middle of the box indicates the second quartile (median). Dots above and below the box are outliers. Plates were interpreted after 18-24 hr (day 1) and 42-48 hr (day 2) of incubation during working days. On working days, the median reporting time was 32.9 hr (range: 19.0-55.2 hr) and 104.1 hr (range: 69.8-165.5 hr). When including holidays and weekends, the median of reporting time was increased to 33.7 hr (range: 18.2-156.9 hr) and 108.1 hr (range: 69.8-181.0 hr). Colonies detected on day 2 required an additional 47.2 hr for subculture and further biochemical tests, which resulted in a delay of reporting of approximately 3 days.
). The bottom and top edges of the boxes mark the first quartile and third quartile, respectively. The horizontal line in the middle of the box indicates the second quartile (median). Dots above and below the box are outliers. Plates were interpreted after 18-24 hr (day 1) and 42-48 hr (day 2) of incubation during working days. On working days, the median reporting time was 32.9 hr (range: 19.0-55.2 hr) and 104.1 hr (range: 69.8-165.5 hr). When including holidays and weekends, the median of reporting time was increased to 33.7 hr (range: 18.2-156.9 hr) and 108.1 hr (range: 69.8-181.0 hr). Colonies detected on day 2 required an additional 47.2 hr for subculture and further biochemical tests, which resulted in a delay of reporting of approximately 3 days.
 ). The bottom and top edges of the boxes mark the first quartile and third quartile, respectively. The horizontal line in the middle of the box indicates the second quartile (median). Dots above and below the box are outliers. Plates were interpreted after 18-24 hr (day 1) and 42-48 hr (day 2) of incubation during working days. On working days, the median reporting time was 32.9 hr (range: 19.0-55.2 hr) and 104.1 hr (range: 69.8-165.5 hr). When including holidays and weekends, the median of reporting time was increased to 33.7 hr (range: 18.2-156.9 hr) and 108.1 hr (range: 69.8-181.0 hr). Colonies detected on day 2 required an additional 47.2 hr for subculture and further biochemical tests, which resulted in a delay of reporting of approximately 3 days.
). The bottom and top edges of the boxes mark the first quartile and third quartile, respectively. The horizontal line in the middle of the box indicates the second quartile (median). Dots above and below the box are outliers. Plates were interpreted after 18-24 hr (day 1) and 42-48 hr (day 2) of incubation during working days. On working days, the median reporting time was 32.9 hr (range: 19.0-55.2 hr) and 104.1 hr (range: 69.8-165.5 hr). When including holidays and weekends, the median of reporting time was increased to 33.7 hr (range: 18.2-156.9 hr) and 108.1 hr (range: 69.8-181.0 hr). Colonies detected on day 2 required an additional 47.2 hr for subculture and further biochemical tests, which resulted in a delay of reporting of approximately 3 days.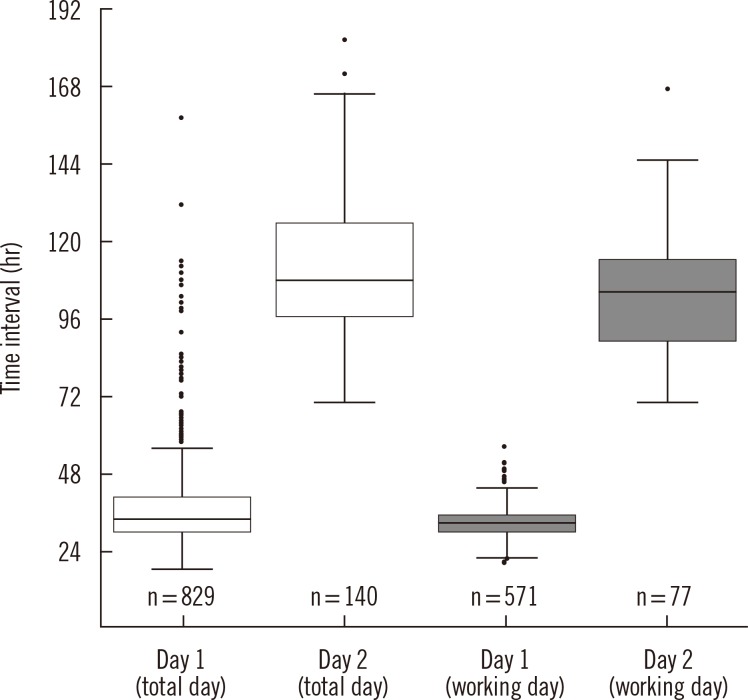




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download