Abstract
Background
Most mutations in the reverse transcriptase (RT) gene of the hepatitis B virus (HBV) are related to resistance to antiviral agents. Cross-sectional studies on the mutations of this gene are rare. Thus, we analyzed the mutation patterns of RT genes and their biochemical parameters.
Methods
From 2009 to 2012, 301 blood specimens from patients with chronic hepatitis B at Daegu Catholic University Medical Center were retrospectively analyzed for the RT gene sequence of HBV, ALT, hepatitis B e antigen (HBeAg), and HBV DNA. The mutation patterns of the RT gene were compared with the biochemical parameters.
Results
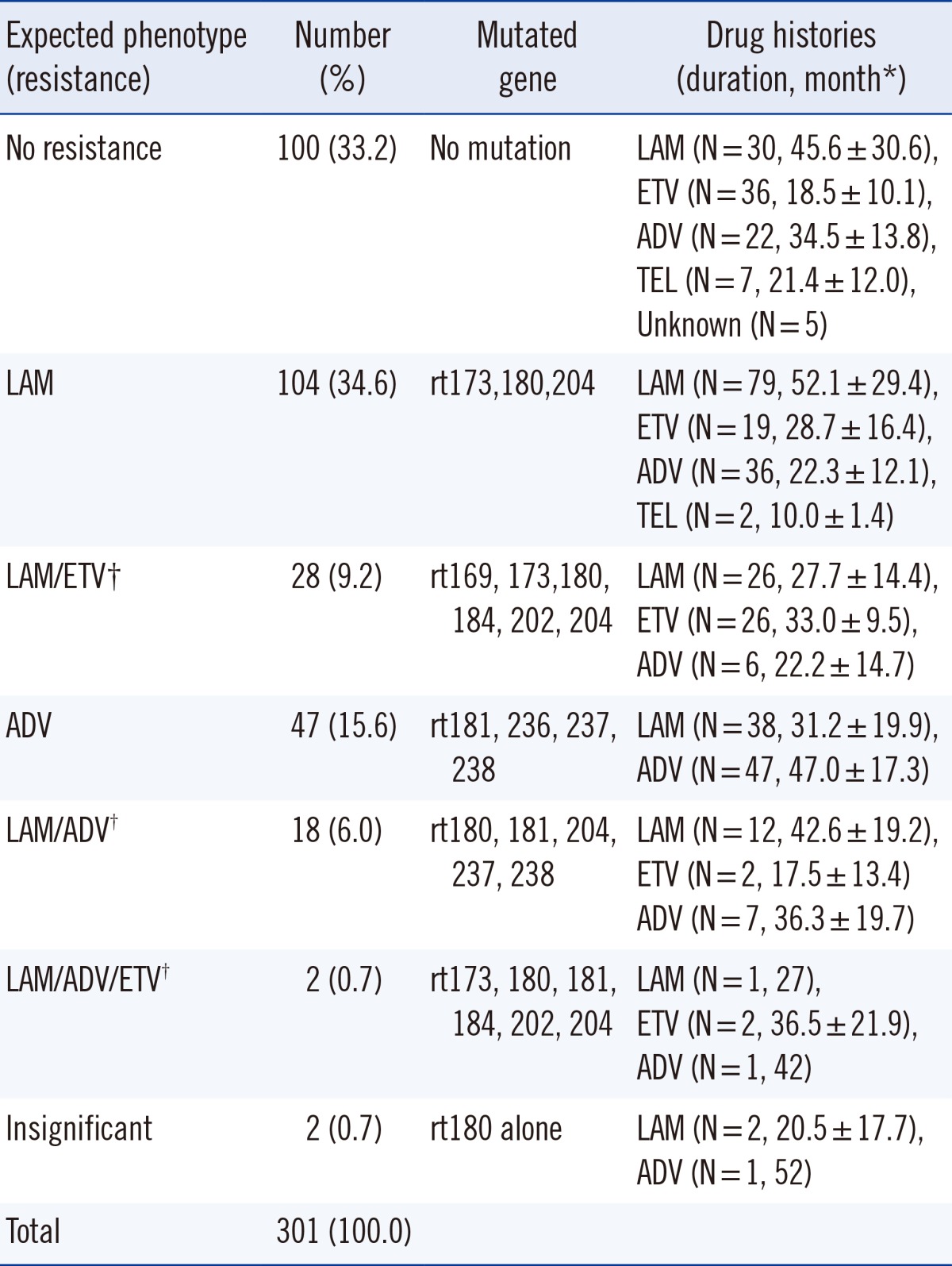
Of the 301 patients, 100 (33.2%) had no RT gene mutations. The remaining showed the following mutation patterns: rtM204I/V (50.2%), rtL180M (39.2%), and rtA181T/V (19.6%). Combined mutations were found in 146 cases (48.5%). Of these, the combination of amino acid changes at rt180+rt204 (49.3%) was most frequently detected, followed by rt181+rt236 (11.0%) and rt173+rt180+rt204 (9.6%). In the mutated group, HBV DNA and HBeAg positive rates were significantly higher (P<0.05 for both). Phenotypic analysis showed that lamivudine resistance was most frequently detected (34.6%), followed by adefovir resistance (15.6%). Multidrug resistance was detected in 48 cases (15.9%). The adefovir-resistant group had a higher proportion of cases with HBV loads greater than 2,000 IU/mL.
The average prevalence of hepatitis B surface antigen (HBsAg) in Korea was 3.2% in 2009 [1], making it an intermediate endemic area. In high endemic countries, the hepatitis B virus (HBV) carrier rate is over 8%, while in low endemic countries, the rate is less than 2% [2].
Mutations in the reverse transcriptase (RT) gene of HBV are closely correlated with resistance to antiviral drugs [3, 4, 5]. The incidence, clinical outcome, and biological significance of these mutants have been studied extensively. However, there have been only a few recent reports about HBV RT mutants in untreated patients. Huang et al. [6] reported a 26.9% tyrosine, methionine, aspartate, aspartate (YMDD) mutation rate from 104 cases who had not received lamivudine or any other antiviral drugs within the last one year. Zhao et al. [7] reported the rate of natural YMDD mutations in Western China as 15.56% from 270 antiviral non-treated (treatment naïve) chronic hepatitis B patients. Moreover, Tan et al. [8] found natural YMDD mutations in 23.3% of 1,042 patients recruited from six centers in China. They posited that the natural existence of YMDD mutant strains was associated with a larger HBV load in chronic hepatitis B patients. However, their report did not rule out the possibility of mutant virus infection from other sources, such as transmission from a carrier having a mutant HBV to treatment naïve chronic hepatitis B patients. Generally, YMDD mutant viruses disappear in a few months after lamivudine withdrawal [9]. However, during lamivudine therapy, patients' continued social activity could be a possible source of HBV infection. Chronic hepatitis B patients infected with HBV mutants usually have the hepatitis B e antigen (HBeAg), and live among others without isolation in Korea.
Thus, it is necessary to know the prevalence of mutant HBV in the hospital-based survey. Most of the studies about RT gene mutations were longitudinal studies carried out after treatment with antiviral medications [10, 11, 12]. We investigated the cross-sectional mutation rates and patterns of the HBV at a university hospital, then compared them with biochemical parameters.
From 2009 to 2012, 301 specimens obtained from chronic hepatitis B patients who had been diagnosed and received antiviral agents at the Daegu Catholic University Medical Center (DCUMC) were sent to Seoul Clinical Laboratory (SCL) (Seoul, Korea) for sequencing of the RT gene of HBV. Chronic hepatitis B was diagnosed as per the guidelines of the Korean Association of the Study of the Liver [13]. In the SCL, the RT gene from rt169 to rt250 was directly sequenced by using the ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
The biochemical parameters of ALT, HBeAg, and HBV DNA concentration were analyzed in the laboratory department of DCUMC. The levels of ALT and HBeAg were measured using Roche modular analytics (Roche Diagnostics, Mannheim, Germany), and the concentration of HBV DNA was measured using the COBAS AmpliPrep/COBAS TaqMan system (Roche Diagnostics). For the point of mutation analysis, three-months-later ALT values, HBeAg, and DNA results were used for the data analysis. ALT values ≤40 IU/L were considered normal. HBV DNA concentrations were measured quantitatively, and we used the percentage of higher DNA concentration (>2,000 IU/mL) for comparison of the selected groups. The mutation patterns were analyzed and compared with the results of the biochemical parameters.
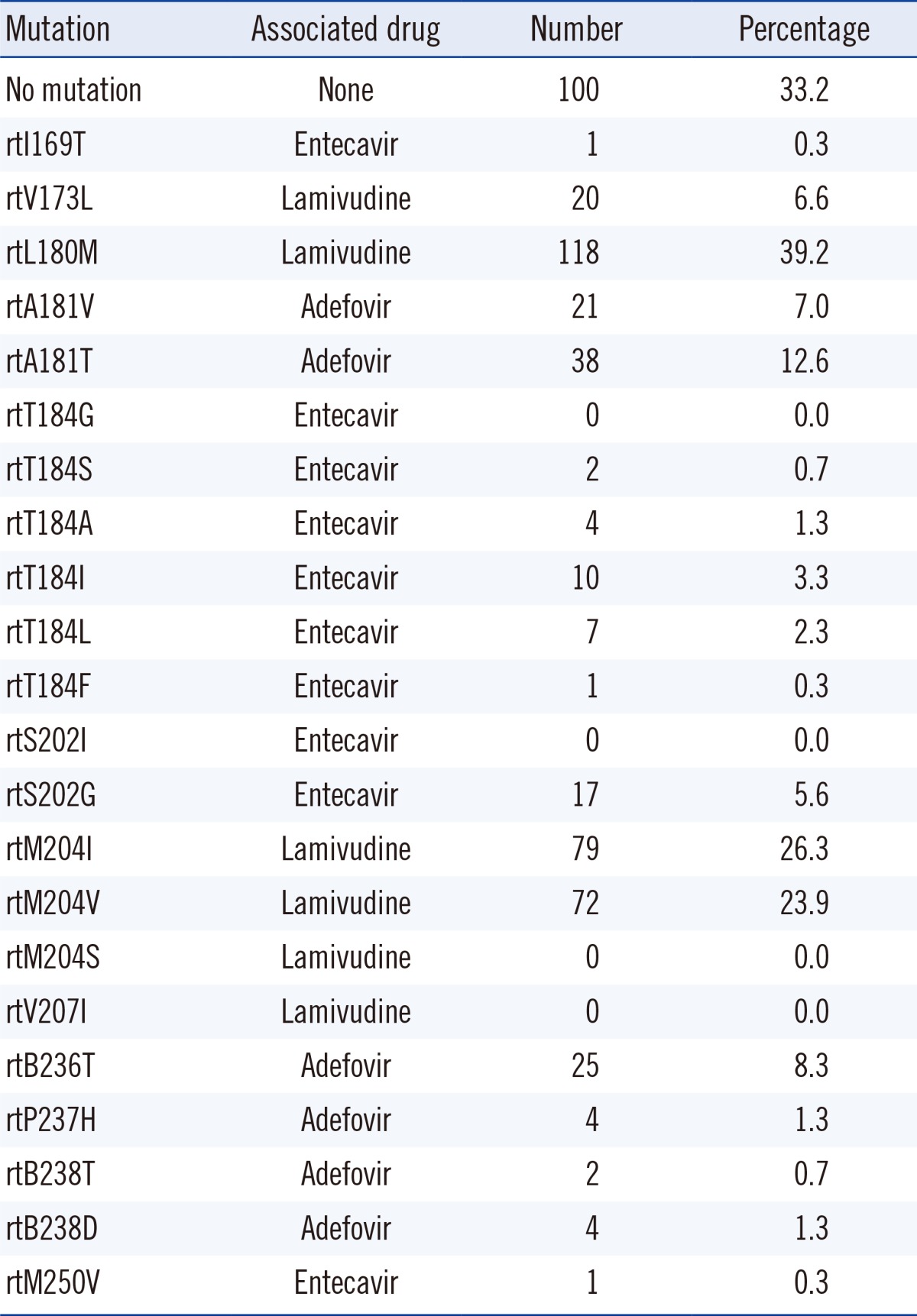
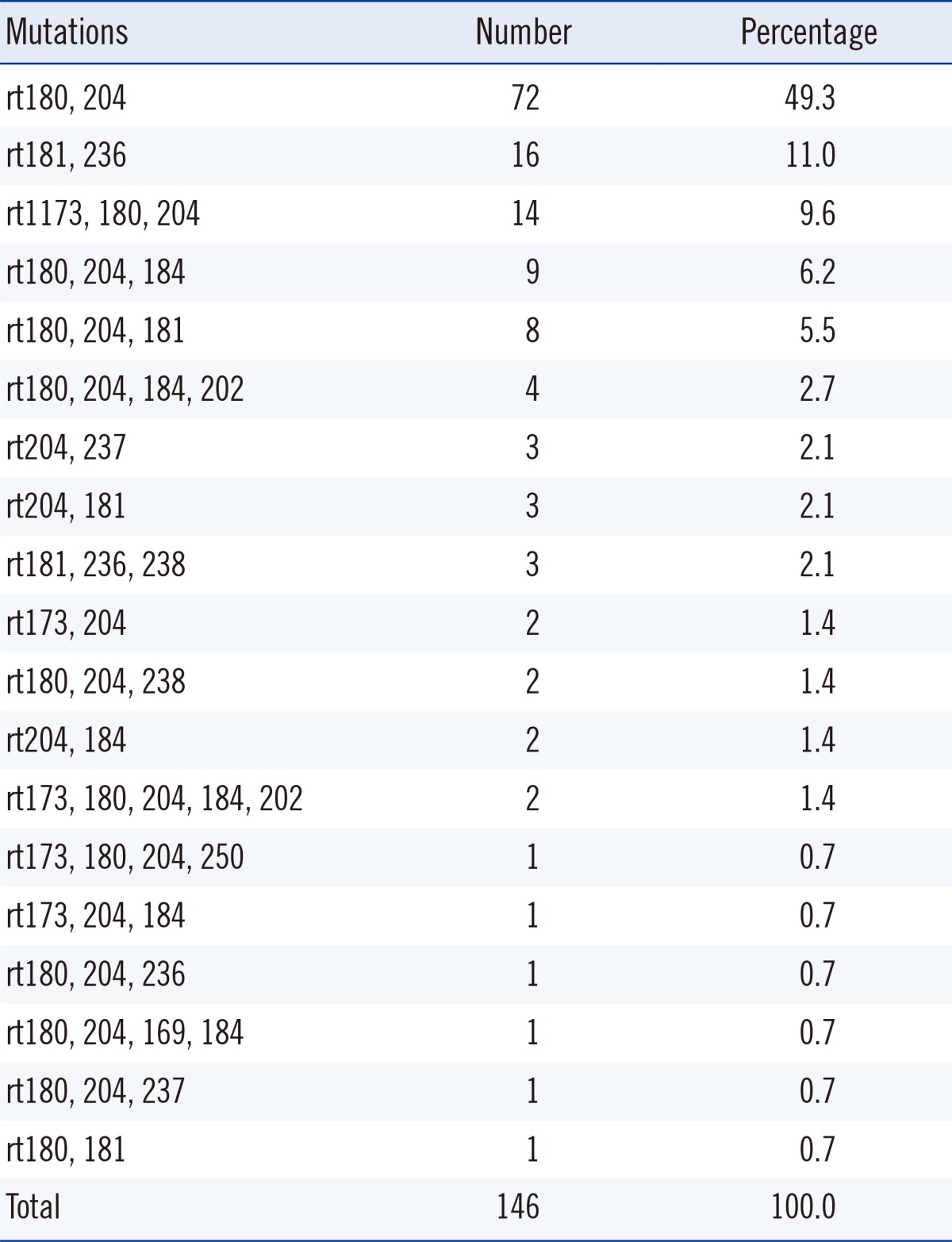
The amino acid changes of the RT gene from 301 specimens are described in Table 1. The rtM204I/V mutation (50.2%) was most frequently detected, followed by rtL180M (39.2%) and rtA181V/T (19.6%) mutations. No mutation was observed in 33.2% of the specimens. The presence of combined mutations was found in 146 cases (48.5%), and the frequency of combined mutations is shown in Table 2. Of the 146 cases, amino acid changes at rt180+rt204 (49.3%) was most frequently detected, followed by rt181+rt236 (11.0%) and rt173+rt180+rt204 (9.6%).
It is difficult to ascertain the clinical significance by mutation analysis only using genotype. Thus, we classified the mutation patterns according to the expected phenotypes associated with drug resistance (Table 3). Lamivudine resistance was most frequently detected (34.6%), followed by adefovir resistance (15.6%). Multidrug resistance was detected in 48 cases (15.9%). The type and duration of drug usage are shown in Table 3.
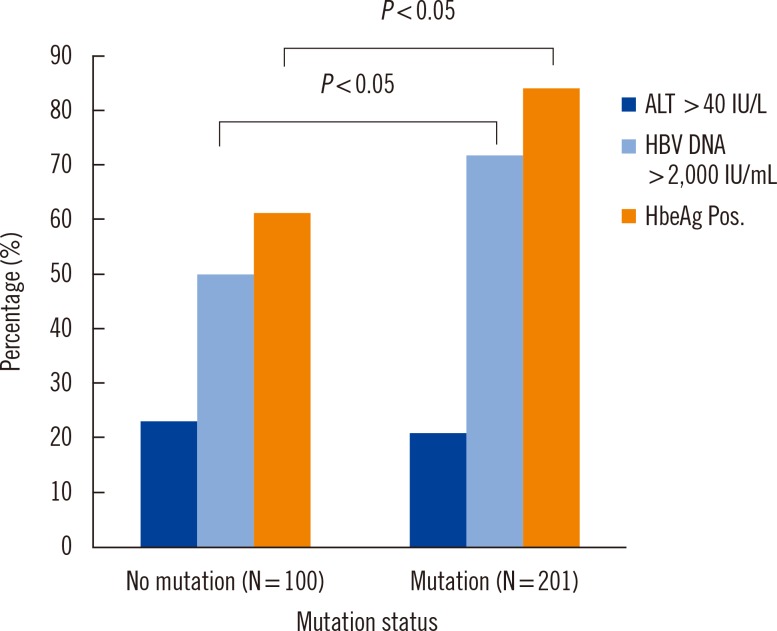
To determine the clinical significance of HBV RT gene mutations, we compared the ALT, HBV DNA concentration, and HBeAg positive rates between mutation-positive and -negative groups. In the mutation-positive group, HBV DNA and HBeAg positive rates were significantly higher (P<0.05, Fig. 1).
When the rtM204V and rtM204I mutation groups were compared, all 72 cases of rtM204V mutation had a simultaneous rtL180M mutation, while in 79 cases of rtM204I mutation, 43 cases (54.4%) had a simultaneous rtL180M mutation, 33 cases (41.8%) had the rtM204I mutation alone, and the remaining 3 cases (3.8%) showed a simultaneous rtV173L mutation. There was no statistical difference of HBeAg positive rates among rtM204V, rtL180M, and rtM204I mutant groups. There were no differences in ALT or HBV DNA levels among the different groups.
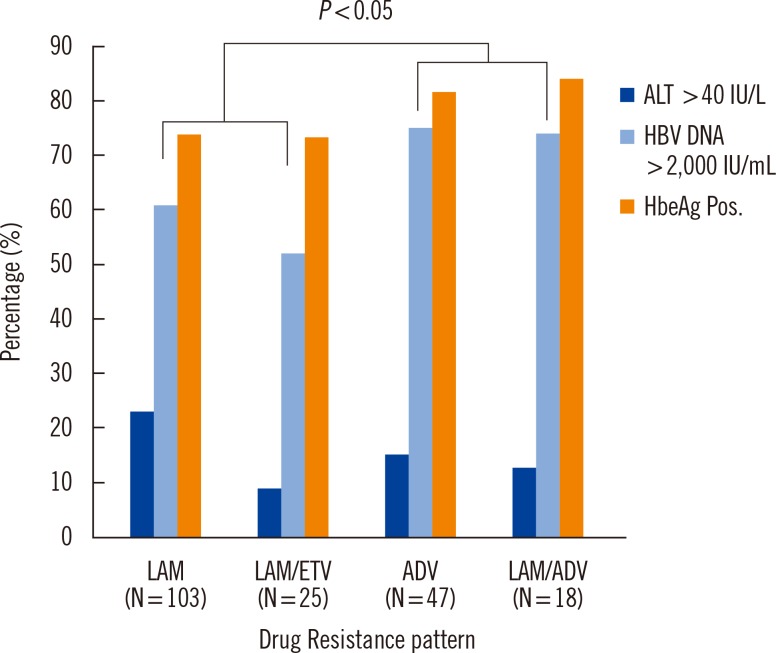
In the drug phenotype analysis of mutation, the adefovir-resistant group showed significantly higher HBV DNA concentration than the lamivudine-resistant group (P<0.05, Fig. 2). The adefovir-resistant group showed higher HBeAg positive rates than the lamivudine-resistant group but the difference was not significant (P=0.087).
Most HBV RT gene mutation analyses are individualized longitudinal studies carried out after the administration of antiviral agents. Important information on drug resistance mechanisms, such as time of appearance of resistance, and medication guidelines for resistant strains have been published and utilized for treatment. However, cross-sectional studies on mutation analysis in admitted patients are very rare. The results of cross-sectional studies might not be as easy to interpret as antimicrobial resistant patterns in bacterial stains. Moreover, genotype to phenotype prediction was difficult owing to the multiple causes of antiviral drug resistance [13]. The therapeutic guidelines for chronic hepatitis B was established in 2004 and subsequently revised in 2007 and in 2011 in Korea [14]. The frequency of RT gene mutations was precipitated by antiviral treatment in Korea. In our study, as expected, the rtM204I/V and rtL180M mutations related to lamivudine resistance were the most frequent, followed by the rtA181V/T mutation associated with adefovir resistance. No mutation was noted in 33.2% of the cases. It was reported that the infection rates of mutant virus in antiviral treatment naïve patients could be influenced by the mutant virus prevalence in the society [7, 8]. Where there is no available data for HBV mutation rates, the mutation rates in the chronic HBV patients visiting hospital might be utilized to postulate the infection rates of mutant virus in antiviral treatment naïve patients.
In the analysis of mutation patterns according to the expected phenotype, multidrug resistance was detected in 15.9% cases. Notably, two cases showing resistance to lamivudine, adefovir, and entecavir were HBeAg positive and had high HBV DNA concentration.
Comparing the mutant and wild type HBV RT groups, the mutant group had significantly higher HBV DNA and HBeAg positive rates. HBV DNA represents the amount of virus present in the blood, and can be used to monitor treatment [15, 16]. HBeAg has been also used to monitor the effectiveness of treatment [15, 17]. Taken together, both markers suggest that the virus is actively replicating and that the infection can be passed to others. Our results suggested that the mutated group had a higher likelihood of infecting healthy people.
The cases with the rtM204I mutation had higher HBeAg positive rates than the cases with rtM204V/rtL180M; however, this difference was statistically insignificant. The rtM204V/rtL180M mutations are associated with much lower resistance to lamivudine than rtM204I alone [18]. In our study, rtM204V was always accompanied by rtL180M. The rtM204I mutation occurs alone followed by rtL180M mutation to increase viral replication [18]. Ono et al. [19] reported that rtM204I mutation alone had lower replication competency than rtL180M/rtM204V, but clinically there was no difference in ALT, HBV DNA levels, and HBeAg positivity [18], similar to our data.
Although genotype to phenotype correlation was difficult, the adefovir-resistant group showed significantly higher HBeAg-positive rates than the lamivudine-resistant group. Since adefovir is usually used to treat lamivudine-resistant cases [5, 14], these results were concordant with the clinical course of chronic hepatitis B patients. Our data showed that HBV replication was more difficult to suppress in the adefovir-resistant group than in the lamivudine-resistant group. We also showed increased HBV DNA concentrations in the adefovir-resistant group.
In conclusion, our results showed some correlation between RT gene mutation status and biochemical parameters. The mutated group showed significantly higher HBV DNA and HBeAg positive rates than the wild type group. The adefovir-resistant group showed significantly higher HBeAg positive rates than the lamivudine-resistant group. The results of RT gene mutations could be utilized to predict clinical status.
References
1. Korea Centers for Disease Control and Prevention. The Fourth Korea National Health and Nutrition Examination Survey (KNHANES IV) 2009-Health Examination. Korea Centers for Disease Control and Prevention;2010.
2. Franco E, Bagnato B, Marino MG, Meleleo C, Serino L, Zaratti L. Hepatitis B: Epidemiology and prevention in developing countries. World J Hepatol. 2012; 4:74–80. PMID: 22489259.

3. Hussain M, Lok AS. Mutations in the hepatitis B virus polymerase gene associated with antiviral treatment for hepatitis B. J Viral Hepat. 1999; 6:183–194. PMID: 10607230.

4. Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol. 2006; 44:593–606. PMID: 16455151.

5. Yim HJ. Hepatitis B virus genetic diversity and mutant. Korean J Hepatol. 2008; 14:446–464. PMID: 19119240.

6. Huang ZM, Huang QW, Qin YQ, He YZ, Qin HJ, Zhou YN, et al. YMDD mutations in patients with chronic hepatitis B untreated with antiviral medicines. World J Gastroenterol. 2005; 11:867–870. PMID: 15682483.

7. Zhao J, Guo Y, Yan Z, Liang P, Zhang J, Liu Y. The natural YMDD mutations of hepatitis B virus in Western China. Scand J Infect Dis. 2012; 44:44–47. PMID: 21851334.

8. Tan YW, Ge GH, Zhao W, Gan JH, Zhao Y, Niu ZL, et al. YMDD motif mutations in chronic hepatitis B antiviral treatment naïve patients: a multi-center study. Braz J Infect Dis. 2012; 16:250–255. PMID: 22729192.

9. Orlando R, Tosone G, Portella G, Veropalumbo E, D'Onofrio M, Piazza M. Prolonged persistence of lamivudine-resistant mutant and emergence of new lamivudine-resistant mutants two years after lamivudine withdrawal in HBsAg-positive chronic hepatitis patient: a case report. Infection. 2008; 36:472–474. PMID: 17962902.

10. Wang F, Wang H, Shen H, Meng C, Weng X, Zhang W. Evolution of hepatitis B virus polymerase mutations in a patient with HBeAg-positive chronic hepatitis B virus treated with sequential monotherapy and add-on nucleoside/nucleotide analogues. Clin Ther. 2009; 31:360–366. PMID: 19302908.

11. Bartholomeusz A, Locarnini S. Hepatitis B virus mutations associated with antiviral therapy. J Med Virol. 2006; 78(S1):S52–S55. PMID: 16622878.

12. Moon JH, Cho M, Yoon KT, Bae JH, Heo J, Kim GH, et al. The efficacy of adefovir dipivoxil monotherapy and the incidence of genotypic resistance to adefovir dipivoxil in patients with lamivudine-resistant chronic hepatitis B infection. Korean J Hepatol. 2008; 14:503–512. PMID: 19119245.

13. Locarnini S, Warner N. Major causes of antiviral drug resistance and implications for treatment of hepatitis B virus monoinfection and coinfection with HIV. Antivir Ther. 2007; 12(S3):H15–H23. PMID: 18284179.
14. Korean Association for the Study of the Liver. KASL Clinical Practice Guidelines: Management of chronic hepatitis B. Clin Mol Hepatol. 2012; 18:109–162. PMID: 22893865.
15. Yuen MF, Ng IO, Fan ST, Yuan HJ, Wong DK, Yuen JC, et al. Significance of HBV DNA levels in liver histology of HBeAg and Anti-HBe positive patients with chronic hepatitis B. Am J Gastroenterol. 2004; 99:2032–2037. PMID: 15447768.

16. Mommeja-Marin H, Mondou E, Blum MR, Rousseau F. Serum HBV DNA as a marker of efficacy during therapy for chronic HBV infection: analysis and review of the literature. Hepatology. 2003; 37:1309–1319. PMID: 12774009.

17. Alagiozian-Angelova V, Alagiozian D, Antonov K, Krustev Z. Clinical significance of serum HBeAg and HBV-DNA-specific values of virus replication in chronic hepatitis-B virus infection. Folia Med (Plovdiv). 1998; 40:34–41. PMID: 10371797.
18. Enomoto M, Tamori A, Kohmoto MT, Morikawa H, Habu D, Sakaguchi H, et al. Mutational patterns of hepatitis B virus genome and clinical outcomes after emergence of drug-resistant variants during lamivudine therapy: analyses of the polymerase gene and full-length sequences. J Med Virol. 2007; 79:1664–1670. PMID: 17854034.

19. Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF, et al. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest. 2001; 107:449–455. PMID: 11181644.

Fig. 1
Comparison of biochemical parameters according to the mutation status of the hepatitis B virus reverse transcriptase gene.
Abbreviations: HBV, hepatitis B virus; HBeAg Pos, hepatitis B e antigen positive.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download