Abstract
Background
Streptococcus pneumoniae causes life-threatening infections such as meningitis, pneumonia, and febrile bacteremia, particularly in young children. The increasing number of drug-resistant isolates has highlighted the necessity for intervening and controlling disease. To achieve this, information is needed on serotype distribution and patterns of antibiotic resistance in children.
Methods
All cases of invasive pneumococcal disease (IPD) in children aged less than 15 yr recorded at King Khalid University Hospital, King Saud University, Riyadh, Saudi Arabia, were reviewed for serotyping and antibiotic susceptibility. Isolates were collected from 78 consecutive patients with IPD between 2009 and 2012. All collected isolates were subjected to serotyping by co-agglutination, sequential multiplex PCR, and single PCR sequetyping as previously described.
Results
The most frequently isolated IPD serotypes were 23F, 6B, 19F, 18C, 4, 14, and 19A, which are listed in decreasing order and cover 77% of total isolates. The serotype coverage for the pneumococcal conjugate vaccine (PCV)7, PCV10, and PCV13 was 77%, 81%, and 90%, respectively. Results from sequential multiplex PCR agreed with co-agglutination results. All serotypes could not be correctly identified using single PCR sequetyping. Minimum inhibitory concentration showed that 50 (64%) isolates were susceptible to penicillin, whereas 70 (90%) isolates were susceptible to cefotaxime.
Conclusions
The most common pneumococcal serotypes occur with frequencies similar to those found in countries where the PCV has been introduced. The most common serotypes in this study are included in the PCVs. Addition of 23A and 15 to the vaccine would improve the PCV performance in IPD prevention.
Streptococcus pneumoniae is a causative agent of many serious community-acquired systemic infections, as well as more localized (upper respiratory tract, pneumonia, and otitis media) infections [1, 2, 3]. It colonizes the upper respiratory tract as a commensal but modulates and transforms into a pathogen that targets infants, elderly, and immune-compromised patients, and causes life-threatening diseases such as pneumonia, bacteremia, and meningitis [3, 4, 5]. The enormous heterogeneity of its polysaccharide capsule, which affects the virulence in this bacterium, creates a major impediment for designing effective vaccines [6]. The antiphagocytic polysaccharide capsule is a major virulence factor of pneumococci, and central regions of the loci contain serotype-specific genes that form the basis of multiplex PCR schemes [7]. On the basis of the immunochemistry of the capsular polysaccharide, S. pneumoniae is classified into more than 93 serotypes [5], of which 15 serotypes (14, 6, 1, 19, 3, 4, 5, 9, 18, 23, 12, 7, 2, 25, and 8) are known to cause approximately 90% of the invasive diseases worldwide [5, 8]. The serotypes have different abilities to cause invasive disease, and some serotypes tend to infect certain risk groups such as children or patients with underlying diseases. Several pneumococcal conjugate vaccines (PCV) have been developed and licensed, including PCV7, PCV10, PCV13, and PCV23 [9, 10]. Because geographical variations in serotype distribution continue to be reported globally, a single vaccine may not be universally optimal. Knowledge of regional pneumococcal serotype distribution is required to infer the potential impact of vaccination in a regional population being immunized.
Quellung, or the co-agglutination method, is the accepted reference method for direct determination of the capsular serotypes, but it has limitations due to cost and required technical expertise [11, 12]. Sequential multiplex PCR (SM-PCR) was developed more recently and has shown the greatest value for monitoring capsular evolution because it is more cost-effective than conventional serotyping of S. pneumoniae [12]. Recently, a single PCR sequencing serotyping method was described on the basis of a computer algorithm to interrogate the capsulation locus (cps) of vaccine serotypes in order to locate primer pairs in conserved regions that border variable regions and can differentiate among serotypes [13]. The present study was undertaken to determine the serotype distribution of invasive S. pneumoniae isolated from children and to correlate this distribution with antibiotic susceptibility.
The study was designed and approved by the institutional ethics committee of King Khalid University Hospital at King Saud University, Riyadh, Saudi Arabia. Blood, cerebrospinal fluid (CSF), pleural, ascites and synovial fluid samples of children under age 15 yr were collected from all suspected cases of pneumonia and meningitis. Samples were processed according to standard operating procedures, and all recovered isolates were identified and confirmed as S. pneumoniae by colony morphology, hemolysis on blood agar plates, optochin sensitivity, and bile solubility testing [1, 2]. If several isolates were extracted from the same patient, only one isolate from the same disease episode was included in the study. Molecular identification was accomplished by targeting the lytA region by using primers as described elsewhere [14]. S. pneumoniae isolates were serotyped by the co-agglutination test as described earlier [15], which can deduce all known capsular serotypes by using the antisera obtained from Staten's Serum Institute, Copenhagen, Denmark.
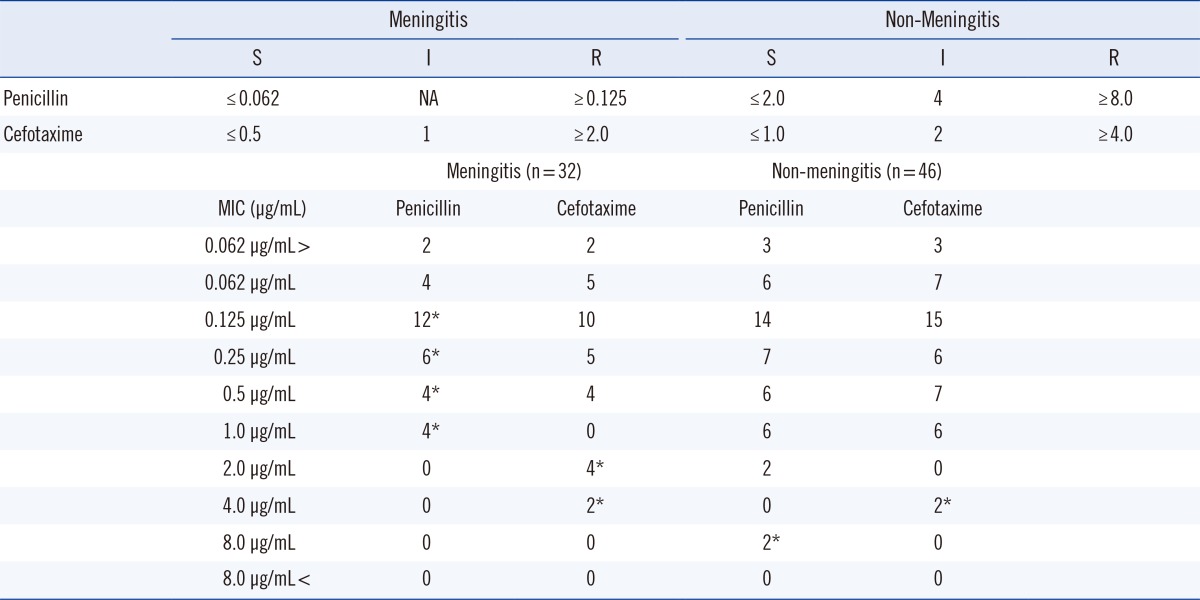
Drug susceptibility testing was performed using disk diffusion on Mueller-Hinton blood agar supplemented with 7% sheep blood agar. In addition, penicillin and cefotaxime minimum inhibitory concentrations (MIC) were determined for all isolates with the E-test according to the manufacturer's recommendations (AB Biodisk, Solna, Sweden). The revised breakpoints of parenteral cut-offs were used according to the CLSI 2009 guidelines [16]. S. pneumonia isolates associated with meningitis, for which the MICs of penicillin and cefotaxime were equal to or more than 0.125 and 2.0 µg/mL, respectively, were considered resistant; the isolates, for which MICs of penicillin and cefotaxime that were equal to or less than 0.062 and 0.5 µg/mL, respectively, were susceptible. In non-meningitis cases, penicillin and cefotaxime MICs equal to or more than 8.0 and 4.0 µg/mL, respectively, were considered resistant; MICs equal to or less than 2.0 and 1.0 µg/mL, respectively, were susceptible (Table 1). S. pneumoniae (ATCC 49619) was used as the reference strain for all the laboratory procedures.
DNA extraction, PCR, and product detection were performed as described earlier [12]. To identify the prevalent serotypes, SM-PCR reactions were run with a total of 40 serotypes and the internal positive control for the cpsA locus. In brief, 40 primers were grouped into seven multiplex reactions. Each reaction was designed to include four primer pairs targeting serotype-specific regions of four different serotypes. Each reaction also included an internal positive control targeting all known pneumococcal cps operons.
A conventional PCR was performed to amplify cpsB target gene by using primers as follows: cps1, 5'-GCA ATG CCA GAC AGT AAC CTC TAT-3' and cps2, 5'-CCT GCC TGC AAG TCT TGA TT-3'. The PCR reaction mixture and number of cycles were as described earlier [13]. PCR amplicons were analyzed with 1.5% agarose gel. The expected cps band sizes of 1,000 bp were purified and sequenced in both directions with the same primers. Sequences were analyzed and interrogated in the GenBank database. Serogroup/types of isolates were defined as described [13].
Isolates that could not be typed by SM-PCR but were positive for the cpsA locus were serotyped by the co-agglutination reaction. For strains with discrepant results among conventional serotyping, SM-PCR, and sequetyping, typing methods were repeated independently.
During the study period, 78 S. pneumoniae isolates were recovered from children aged less than 15 yr. The isolates included 46 (59%) cases of pneumonia and 32 (41%) cases of meningitis. The condition was diagnosed as meningitis on the basis of increased CSF pressure, a high cell count (108-1010/L), increased protein (>100 mg/dL), and decreased glucose (<40 mg/dL or <50% of the simultaneous glucose blood level). Sixty-nine (88%) invasive isolates were recovered from blood, 6 (8%) from CSF, and 3 (4%) from other sterile fluids, one each from peritoneal, synovial, and pleural fluid. Approximately 40% were isolated from children aged less than 5 yr.
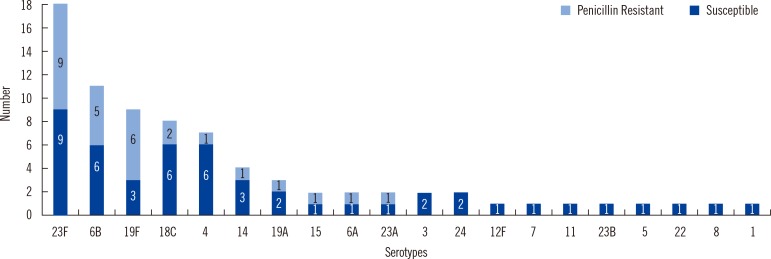
Seventy isolates (90%) were found to be resistant to penicillin, as determined from the results of the 1-µg oxacillin disk-diffusion test. However, MIC values showed that 50 (64%) isolates were susceptible to penicillin, and 70 (90%) isolates were susceptible to cefotaxime. MICs for penicillin ranged between 0.062 and 1.0 µg/mL and 0.062 and 8.0 µg/mL for meningitis- and non-meningitis-associated cases, respectively (Table 1). A total of 60 (77%) isolates were erythromycin-resistant, and all 78 (100%) were resistant to co-trimoxazole (Fig. 1). All the isolates were susceptible to vancomycin, linezolid, and levofloxacin. Resistance to penicillin was observed in a higher number of serotypes 23F, 6B, and 19 F than in the other serotypes (Fig. 2).
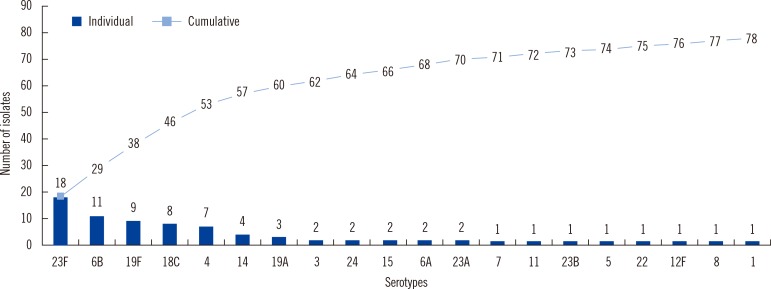
The serotype coverage for the pneumococcal vaccines PCV7, PCV10, and PCV13 was 77%, 81%, and 90%, respectively. The most frequently isolated IPD serotypes were 23F, 6B, 19F, 18C, 4, 14, and 19A, listed in decreasing order; they were found in 77% of the isolates (Fig. 3 and 4).
All the isolates were successfully typed by SM-PCR, and the results were 100% consistent with those of co-agglutination. All serotypes could not be correctly identified using single PCR sequetyping. Of the 78 strains tested, 61 (78%) were sequence-typed to the correct serotype. An additional 14 (18%) strains were sequence-typed to the serogroup level, and 3 (4%) gave vague results. Among nine strains of serotype 19F, one was identified as serotype 1 with low sequence identity (96%), 12F was identified as 12B, and 18B/C could not be differentiated as individual serotypes by sequetyping.
S. pneumoniae is one of the most common causes of morbidity and mortality worldwide, especially in children, and causes life-threatening infections such as meningitis, pneumonia, and febrile bacteremia. S. pneumoniae is a key factor in the development of invasive disease and the spread of resistant strains within the community. Emergence of drug-resistant isolates has highlighted the need for prevention of invasive pneumococcal disease (IPD) as the best method for controlling disease. To achieve this, detailed information is needed for determining the serotype distribution and patterns of antibiotic resistance [3, 5, 10, 11].
The most frequent pneumococcal serotypes were 23F, 6B, 19F, 14, 18C, and 4 (Fig. 4), which covers 73% of total serotypes and six of the seven serotypes in the seven-valent conjugate vaccine (PCV7); serotype 9V was less common. Each of serotypes 1 and 5 was isolated once. The serotype coverage for the PCV7, PCV10, and PCV13 was 77%, 81%, and 90%, respectively (Fig. 3). The proportion of IPD caused by vaccine types was higher compared to previous similar studies [17, 18, 19, 20]. The serotype distribution in the present investigation was similar to that found in previous studies [17, 21], except that serotype 14 was less prevalent than that found in an earlier study [20]. Overall, the isolated serotypes included the most common serotypes causing invasive disease, with most being represented in PCV7. Non-PCV13 vaccine serotypes are 10% of the total. Because the sample size is small, there is a need to do surveillance on large populations and isolates. Therefore, 13 serotypes would include 90% of invasive isolates.
All the isolates were resistant to co-trimoxazole, whereas 77% and 36% resistance was observed against erythromycin and penicillin, respectively. Resistance to cefotaxime and chloramphenicol was far less. The results are very similar to the results from Saudi Arabia and other parts of the world [17, 20, 21]. Isolates were highly susceptible to vancomycin, linezolid, and levofloxacin.
We also evaluated the specificity of SM-PCR and concluded that it is highly specific and sensitive for the identification of the serotype. In our study, PCR results for serogroups of isolates correctly matched those of co-agglutination, while co-agglutination identifies complete serotypes for all isolates. The major disadvantage of the current SM-PCR is that it does not type an isolate completely, especially the commonly occurring 6A/6B/6C serotypes of group 6, 12F/A serotypes of group 12, 15B/C serotypes of group 15, 7F and 7A, 9V and 9A, 33F, 33A and 37, 35A, 35C, and 42. The sequence typing method has big advantage over other techniques, as it requires only a single PCR amplification. It is straight forward, robust, and requires only crude genomic DNA from heat-lysed cells. In addition, sequencing facilities can serotype without the need for an expensive set of serological reagents. However, these results are based on only 20 different serotypes. A more detailed evaluation of sequence typing with a greater number of different serotypes will provide a better evaluation of this scheme.
SM-PCR reduces the time consumed for assessing strains that need to be completely serotyped by conventional co-agglutination techniques. The advantages that SM-PCR has over the other two serotyping methods are that it is a simple, fast, and reliable method, interpretation of the results is not subjective, and it does not require highly specialized expertise.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project no. RGP-VPP-314.
References
1. Richter SS, Heilmann KP, Dohrn CL, Riahi F, Beekmann SE, Doern GV. Accuracy of phenotypic methods for identification of Streptococcus pneumoniae isolates included in surveillance programs. J Clin Microbiol. 2008; 46:2184–2188. PMID: 18495854.
2. Arbique JC, Poyart C, Trieu-Cuot P, Quesne G, Carvalho Mda G, Steigerwalt AG, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004; 42:4686–4696. PMID: 15472328.
3. James J, Gnanapragasam HP, Sangeetha G. Causes and epidemiology of vaccine preventable infectious bacterial disease: The prospect and short out coming of vaccine. Int J Pharm Pharm Sci. 2012; 4:51–54.
4. Dias CA, Teixeira LM, Carvalho Mda G, Beall B. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J Med Microbiol. 2007; 56:1185–1188. PMID: 17761481.

5. Lee JH, Kim SH, Lee J, Choi EH, Lee HJ. Diagnosis of pneumococcal empyema using immunochromatographic test on pleural fluid and serotype distribution in Korean children. Diagn Microbiol Infect Dis. 2012; 72:119–124. PMID: 22079140.

6. Hausdorff WP, Bryant J, Paradiso PR, Siber GR. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis. 2000; 30:100–121. PMID: 10619740.

7. Park IH, Park S, Hollingshead SK, Nahm MH. Genetic basis for the new pneumococcal serotype, 6C. Infect Immun. 2007; 75:4482–4489. PMID: 17576753.

8. Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, et al. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol. 2013; 51:647–652. PMID: 23224094.

9. Paradiso PR. Advances in pneumococcal disease prevention: 13-valent pneumococcal conjugate vaccine for infants and children. Clin Infect Dis. 2011; 52:1241–1247. PMID: 21507921.

10. Fedson DS, Guppy MJ. Pneumococcal vaccination of older adults: conjugate or polysaccharide? Hum Vaccin Immunother. 2013; 9:1382–1384. PMID: 23732892.
11. Brito DA, Ramirez M, de Lencastre H. Serotyping Streptococcus pneumoniae by multiplex PCR. J Clin Microbiol. 2003; 41:2378–2384. PMID: 12791852.
12. Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006; 44:124–131. PMID: 16390959.

13. Leung MH, Bryson K, Freystatter K, Pichon B, Edwards G, Charalambous BM, et al. Sequetyping: serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J Clin Microbiol. 2012; 50:2419–2427. PMID: 22553238.
14. Greve T, Møller JK. Accuracy of using the lytA gene to distinguish Streptococcus pneumoniae from related species. J Med Microbiol. 2012; 61:478–482. PMID: 22135022.
15. Lalitha MK, Thomas K, Kumar RS, Steinhoff MC. the IBIS Study Group. Serotyping of Streptococcus pneumoniae by coagglutination with 12 pooled antisera. J Clin Microbiol. 1999; 37:263–265. PMID: 9854110.
16. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 29th Informational supplement, M100-S19. Wayne, PA: Clinical and Laboratory Standards Institute;2009.
17. Shibl AM, Memish ZA, Al-Kattan KM. Antibiotic resistance and serotype distribution of invasive pneumococcal diseases before and after introduction of pneumococcal conjugate vaccine in the Kingdom of Saudi Arabia (KSA). Vaccine. 2012; 30(S6):G32–G36. PMID: 23228355.

18. Dagan R. Impact of pneumococcal conjugate vaccine on infections caused by antibiotic-resistant Streptococcus pneumoniae. Clin Microbiol Infect. 2009; 15(S3):16–20. PMID: 19366365.
19. Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, et al. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J Infect Dis. 2007; 196:1346–1354. PMID: 17922399.

20. Al-Mazrou A, Twum-Danso K, Al Zamil F, Kambal A. Streptococcus pneumoniae serotypes/serogroups causing invasive disease in Riyadh, Saudi Arabia: extent of coverage by pneumococcal vaccines. Ann Saudi Med. 2005; 25:94–99. PMID: 15977684.
21. Fouda SI, Kadry AA, Shibl AM. Beta-lactam and macrolide resistance and serotype distribution among Streptococcus pneumoniae isolates from Saudi Arabia. J Chemother. 2004; 16:517–523. PMID: 15700841.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download