Abstract
Background
Conventional acid-fast bacilli (AFB) staining cannot differentiate viable from dead cells. Propidium monoazide (PMA) is a photoreactive DNA-binding dye that inhibits PCR amplification by DNA modification. We evaluated whether PMA real-time PCR is suitable for the early detection of viable Mycobacterium tuberculosis (MTB) in clinical respiratory specimens.
Methods
A total of 15 diluted suspensions from 5 clinical MTB isolates were quadruplicated and subjected to PMA treatment and/or heat inactivation. Eighty-three AFB-positive sputum samples were also tested to compare the ΔCT values (CT value in PMA-treated sputum samples-CT value in non-PMA-treated sputum samples) between culture-positive and culture-negative specimens. Real-time PCR was performed using Anyplex MTB/NTM Real-Time Detection (Seegene, Korea), and the CT value changes after PMA treatment were compared between culture-positive and culture-negative groups.
Results
In MTB suspensions, the increase in the CT value after PMA treatment was significant in dead cells (P=0.0001) but not in live cells (P=0.1070). In 14 culture-negative sputum samples, the median ΔCT value was 5.3 (95% confidence interval [CI], 4.1-8.2; P<0.0001), whereas that in 69 culture-positive sputum samples was 1.1 (95% CI, 0.7-2.0). In the ROC curve analysis, the cutoff ΔCT value for maximum sensitivity (89.9%) and specificity (85.7%) for differentiating dead from live cells was 3.4.
Early detection of Mycobacterium tuberculosis (MTB) in clinical specimens is important for the treatment and monitoring of tuberculosis (TB) [1]. Traditionally, the acid-fast bacilli (AFB) smear has been used for the diagnosis and monitoring of TB because of its rapidity and easy applicability. However, it has low sensitivity compared with culture tests and cannot discriminate MTB from non-tuberculous mycobacteria (NTM). It also cannot differentiate viable from dead cells [2].
To improve its sensitivity and specificity, various molecular methods based on nucleic acid amplification have been developed and applied in clinical laboratories [3, 4]. However, none of these approaches can discriminate between viable and dead cells because bacterial DNA (the target of the test) is present in both cases. This renders the methods inappropriate for the monitoring of pulmonary TB during or after treatment [5]. Many AFB smear-positive and culture-negative cases are observed in patients with far-advanced cavitary TB. It is thought that, in many cases, these organisms are dead and that their presence is not a sign of treatment failure [6]. However, if they are still alive, the patients are exposed to the risk of being treated with ineffective regimens until culture results are reported.
The ability to determine the viability of AFB found in smear-positive sputum specimens would improve the monitoring of patients and assist decisions regarding the clinical course. Propidium monoazide (PMA; Biotium, Hayward, CA, USA) is a photoreactive DNA-binding dye that inhibits PCR amplification by DNA modification. This compound can penetrate damaged cell walls but does not permeate intact cell walls; thus, PMA can selectively modify DNA in damaged cells [7]. When dead cells are treated with PMA, the compound inhibits the amplification of DNA so that no PCR amplification signal will be detected or the CT value (a measure of the number of PCR cycles before a specific product is detected) will be higher than that of dead cells not treated with PMA. In viable cells, PMA does not inhibit PCR; therefore, little or no difference would be observed in CT values in the presence of this reagent. We evaluated whether this method could be combined with real-time PCR in order to distinguish living from dead bacteria in clinical respiratory specimens.
This research was reviewed and approved after full committee review by the Institutional Review Board at Pusan National University Yangsan Hospital (PNUYH) (No. 05-2012-065).
The effect of PMA is influenced by its final concentration and the intensity of light exposure. We determined the optimal conditions for PMA treatment for a MTB suspension with a concentration of 2+ according to the WHO guidelines for AFB smear positivity [8]. The 0.5 McFarland suspensions were diluted 1:100. The concentrations of cells in the diluted suspensions were confirmed by smear microscopy with Ziehl-Neelson AFB staining. Cell suspensions were prepared in triplicate in 250-µL volumes for the PMA-untreated control, PMA-treated living cells, and PMA-treated heat-inactivated dead cells. For heat killing, aliquots were heated to 80℃ for 20 min. The tested PMA concentrations were 50 and 100 µM. The light intensities tested ranged from 600 to 800 Watt and the exposure time ranged from 2 to 5 min at a distance of 20 cm from the light source.
DNA extraction and real-time PCR were performed with PMA-treated and PMA-untreated samples, and CT values were calculated. The conditions that induced the maximum increase in the CT value in PMA-treated heat-killed cells compared with PMA-untreated control cells and that induced the least increase in the CT value in PMA-treated live cells compared with PMA-untreated control cells were chosen. The combination of 50 µM PMA concentration, 600 Watt of light intensity, 20 cm of distance from the light source, and 5 min of light exposure was the most appropriate (data not shown). The PMA treatment procedures and real-time PCR process are described below.
Twenty micromoles of PMA stock solution was added to 250-µL aliquots at a concentration of 50 µM. The PMA-mixed samples were incubated for 10 min at room temperature on a rocker (30 rpm) covered with aluminum foil. For 600 W of photoreaction, a combination of two halogen light sources (PluslineR7s, 300 W; Philips, Roosendaal, Netherlands) was used. Samples were laid on an ice block on the rocker set 20 cm from the light source and were exposed to the light for 5 min.
The PMA-treated and PMA-untreated samples were processed identically for DNA extraction and real-time PCR. Cells were harvested by centrifugation at 18,000 g for 15 min and washed in phosphate-buffered saline. Suspensions were centrifuged at 18,000 g for 10 min. Pellets were washed with 1 mL of distilled water, resuspended in 250 µL of nuclease-free water, boiled for 30 min, and finally centrifuged at 18,000 g for 3 min. The supernatant liquid was used for the DNA template. Samples were processed according to the manufacturer's instructions for Anyplex MTB/NTM Real-Time Detection (Seegene, Seoul, Korea), and the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) was used for real-time PCR with 5 µL of DNA template, and CT values were calculated. The real-time PCR device we used generates the result of "not detected" when the CT value reaches 40. If a CT value was close to the reaction end-point in a PMA-untreated specimen and a CT value was not obtained in a PMA-treated specimen, the CT value of the PMA-treated specimen was considered to be the end-point of 40.
A total of five MTB isolates cultured on 3% Ogawa media (Asan pharm, Seoul, Korea) in the mycobacteriology laboratory at PNUYH were selected randomly. Each strain was prepared in 3 concentrations corresponding to scanty, 1+, and 2+, as specified by the WHO guidelines for AFB smear positivity [8]. Briefly, cultured MTB colonies were harvested, and suspensions of 0.5 McFarland turbidity were prepared with distilled water. These suspensions were diluted 1:100, 1:200, and 1:400. The cell concentrations of diluted suspensions were confirmed by smear microscopy with Ziehl-Neelson AFB staining. If the grade of positivity in AFB smears of diluted suspensions was higher than we expected, the dilution factor was increased. Samples were quadruplicated and allocated into live (A) and heat-inactivated (B) groups with or without PMA treatment.
The volume of each sample was adjusted to 250 µL and subjected to PMA treatment procedures as described above. A total of 60 samples were subjected to real-time PCR.
The study included 83 sputum samples from adults (21 women, 62 men) who visited PNUYH and National Masan Hospital (NMH) from May to December 2012 and were undergoing treatment for known pulmonary TB diagnosed by AFB culture. These patients also provided further AFB smears and cultures. The median age of patients was 49.2 yr and ranged from 25 to 76 yr. All the patients were HIV negative. The smears and cultures were processed according to routine laboratory practices. Among them, 36 patients had drug-sensitive TB and 47 patients had multidrug-resistant TB. Briefly, sputum was decontaminated with N-acetyl-L-cysteine (NAC)-2% sodium hydroxide (NaOH). Microscopy was performed following fluorescence staining at PNUYH or Ziehl-Neelson staining at NMH, and the grade of positivity in AFB smears was described as scanty, 1+, 2+, or 3+ according to the WHO guidelines [8]. Smears with positivity from scanty to 2+ were selected for the current study. All sputum samples were cultured using a Mycobacterium Growth Indicator Tube (MGIT) with the BACTEC MGIT 960 system (Becton Dickinson Microbiology Systems, Sparks, MD, USA) and 3% Ogawa media. Culture results were observed prospectively and interpreted as follows: A culture set with MGIT and Ogawa media was considered positive, if MTB growth was observed in one or both media. A culture set was considered negative, if MTB growth was not observed in either medium. Culture sets that had contamination in both media or those in which MTB growth was absent in one medium and contamination was present in the other were excluded from data analysis (Table 1).
Preparation of sputum samples for PMA treatment was undertaken as follows. Five hundred microliters of previously decontaminated sputum samples was used to prepare a pellet by centrifugation at 18,000 g for 15 min followed by washing twice with 1 mL of distilled water. Next, 500 µL of distilled water was added, and the mixture was divided into two parts of 250 µL each, one of which was used for the PMA-treated test sample and the other for the PMA-untreated control. All tests, including real-time PCR, were performed on the same day as the conventional smear and culture examinations.
The PMA-treated and -untreated samples were paired within each of the live (A) groups and heat-inactivated (B) groups for calculating the difference or increase in CT values (ΔCT; CT value of PMA-treated sample-CT value of PMA-untreated sample). The paired data were analyzed using the Wilcoxon signed rank test. The PMA treatment conditions for the pilot study were regarded as valid for differentiating viable from dead cells, if the ΔCT was significant in the heat-inactivated (B) group (P<0.05), whereas that in the live (A) group was not (P≥0.05).
The CT values were obtained from each pair of samples with and without PMA treatment, and ΔCT values were calculated. The ΔCT values were compared with the MTB culture results, and ROC curve analysis was performed to determine the optimal cutoff ΔCT value to differentiate dead from live cells. Sensitivity and specificity were defined as the ability to detect viable cells and to exclude dead cells in the clinical respiratory specimens, respectively. Mann-Whitney test was used to compare ΔCT values between culture-positive and culture-negative specimens. Fisher's exact test was used to assess the association between ΔCT and smear-negative conversion. The statistical analysis was performed with MedCalc statistical program (MedCalc Software, Mariakerke, Belgium).
The medical records of 14 patients who had smear-positive and culture-negative results were reviewed to estimate the relationship between the ΔCT and time to smear-negative conversion. Specifically, the ΔCT values were compared between patients with smear-negative conversion within 1 month and longer than 2 months from the time of the PMA real-time PCR assay. There was no patient whose smear result converted to negative between 1 and 2 months.
Compared with the results for the PMA-untreated live (A) group, the increases in CT values in the PMA-treated live (A) group ranged from -1.5 to 4.9 and were not significant (P=0.107). Compared with the results for the PMA-untreated heat inactivated (B) group, the increase in CT values in the PMA-treated heat-inactivated (B) group ranged from -0.8 to 17.7 and were significant (P=0.0001; Fig. 1). Therefore, we concluded that the conditions of PMA treatment were sufficient to differentiate viable from dead cells.
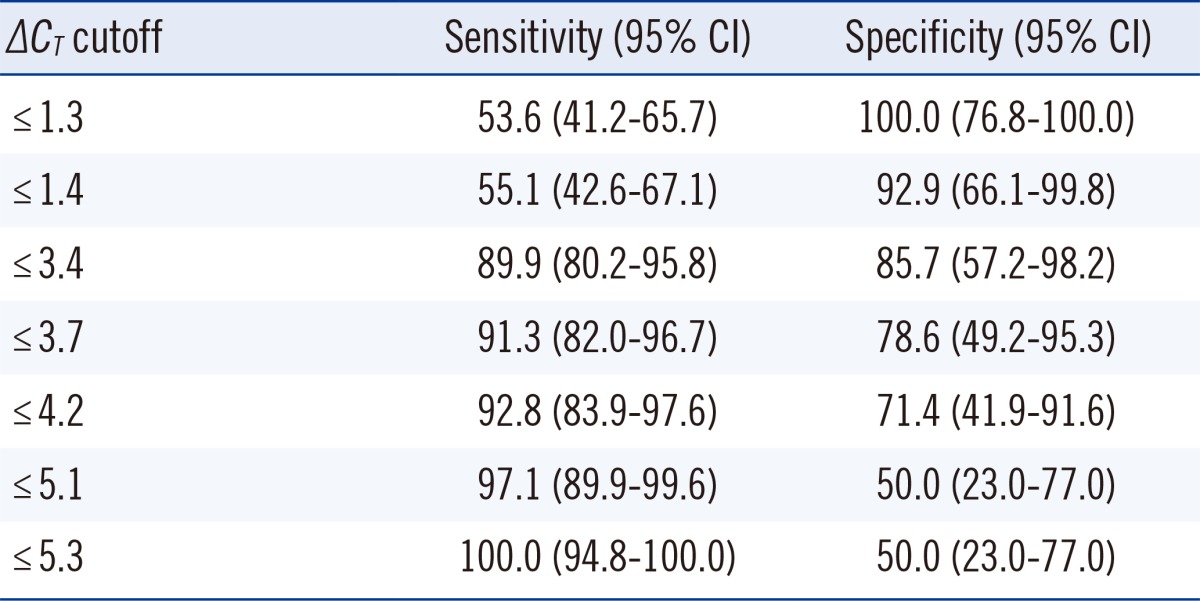
A total of 83 clinical samples (69 MTB culture positive and 14 culture negative) were included for statistical analysis (Table 2). In the culture-positive specimens, the range of ΔCT values was 2.1 to 5.3 and the median value was 1.1 (95% confidence interval [CI], 0.7-2.0). In the culture-negative specimens, ΔCT values ranged from 1.4 to 14.5 and the median value was 5.3 (95% CI, 4.1-8.2), values that were significantly higher than those in culture-positive specimens (P<0.0001). The ROC curves of ΔCT values indicating the sensitivity and specificity for detecting dead bacilli from the smear are depicted in Fig. 2. For these data, the MTB culture results were taken as the reference to indicate viable or dead cells. The area under the curve (AUC) was 0.936. The cutoff ΔCT value for maximum sensitivity (89.9%) and specificity (85.7%) in determining live cells was 3.4. The sensitivity and specificity of various cutoff ΔCT values and 95% CI are shown in Table 3.
Among the 14 patients, one died 2 weeks after the initial AFB smear examination. The remaining 13 patients survived until the end of the study period, and the anti-TB treatment regimens were continued without any change. All 13 patients attained smear-negative conversion. Seven patients for whom negative results were obtained for the smear within 1 month from the time of the PMA real-time PCR assay had ΔCT values above 4.9. Among the 6 patients whose smear results turned negative after 2 or more months, 5 had a ΔCT value below 4.9. Therefore, a ΔCT value above 4.9 was significantly related to smear conversion within a month (P<0.005; Table 4).
The AFB smear and MTB culture are the most important tests for the diagnosis of TB and evaluation of treatment response. A smear produces rapid results within a day; however, it cannot determine the viability of the detected cells. Smear-positive but culture-negative specimens are not common in clinical laboratories. However, when they are encountered, clinical decision-making is delayed until culture results are reported in the case of patients who are undergoing treatment for TB [9]. If the viability of cells in an AFB smear could instead be predicted within a day, rapid action would be possible. The PMA method has been used for rapid differentiation of viable and dead cells for bacterial and fungal strains [10, 11, 12, 13]. The MTB in sputum could also be assessed its viability rapidly by PMA method. Previous studies using PMA and Mycobacterium for predicting viable and dead cells used different methods for PMA treatment than those we used here [13, 14]. The parameters of PMA treatment are influenced by the species, the cell concentrations, and the real-time PCR kit used for detection. The final concentration of PMA in previous studies of MTB ranged from 50 to 500 µM [13, 15]. In our preliminary study described above, 50 µM PMA raised the CT values of samples of smear grade 2+, but not in 3+ smears, while concentrations over 100 µM PMA raised the CT values in samples of viable cells with smear grade 2+ (data not shown). According to annual laboratory statistics from NMH (limited to smear grade 2+), approximately 17.7% of smear-positive cultures were culture-negative [16]. However, culture-negative cases with smear positive grades of 3+ were rarely seen. Therefore, we included specimens of 2+ or lower grades and used 50 µM PMA in our pilot study. The CT values were sufficiently higher for dead cells than for viable cells, and the concentration of 50 µM was adopted for the current study with clinical specimens. A higher concentration of PMA not only induces higher CT values in dead cells but also inhibits the PCR for live cells because of the excessive cell-wall penetration or residual photoactivated PMA in washed samples [15].
For photoactivation of PMA, 500-800 W of light intensity is usually employed [10, 11]. Halogen light can be sufficiently intense to activate PMA, but it causes a rise in temperature. As a result, PMA may be taken up by thermally damaged cells. We exposed the samples to light on an ice block with continuous shaking to minimize the rise in the temperature. By trial and error, we found the best condition for photoactivation of PMA with our subjects to be 600 W, a light-to-sample distance of 20 cm, and 5 min of exposure time. Because of the DNA-binding affinity of photoactivated PMA, residual PMA should be removed before the DNA extraction step to avoid binding to DNA extracted from viable cells. In our initial trial of PCR with unwashed samples after 50 µM PMA treatment, PCR was inhibited, and the signal of the internal control was not observed (data not shown). Furthermore, repeated washing to remove residual PMA can cause loss of cells. We applied the same number of washing steps to PMA-treated and PMA-untreated samples to minimize the gap caused by cell loss in the washing step.
Another point to consider for PMA treatment is that the diverse mechanisms of action of antibiotics may affect the PMA permeability of dead mycobacteria [15]. In our study, the ΔCT values ranged from 1.4 to 14.5 in the culture-negative specimens, a range wider than that reported in a previous study [13]. Usually, most patients with smear-positive and culture-negative results receive treatment. For application of this method to a situation, in which most patients have drug-resistant infections such as NMH, the protocol of PMA treatment and light exposure should be modified and the range of ΔCT values should be evaluated depending on the specific study group. The suggested cutoff ΔCT value for the best sensitivity (89.9%) and specificity (85.7%) in our study was 3.4. However, culture-positive cases with ΔCT above the cutoff value were observed. This can occur because of a mixture of live and dead cells in the specimen [17], for example, in patients receiving treatment or after sputum decontamination. Culture-negative cases with ΔCT values below the cutoff were also observed. In the current study, we evaluated the relation between these low ΔCT values and positive cultures, since we considered positive cultures as evidence of the presence of live bacilli. Strictly speaking, however, not all the viable cells are culturable [18]. Clinical specimens may contain viable but non-culturable bacilli, and those specimens may yield low ΔCT values despite providing negative culture results. This is another limitation of our study.
We noted 2 unexpected observations. First, one sample each in the pilot and the clinical studies had very low ΔCT values. In those cases, the CT values of the PMA-untreated specimens were 38.9 and 38.6, respectively, and CT values from PMA-treated dead cells were not obtained (Fig. 1 and Table 2). The real-time PCR device we used generates the result of "not detected" when the CT value reaches 40. Therefore, when the CT value of the control sample is close to 40, theoretically, the CT value of a PMA-treated dead cell sample cannot reach a value greater than this. This could well be the case in a clinical laboratory, meaning that the ΔCT cannot discriminate live from dead cells in all clinical specimens; this limits the interpretation of the difference in CT values as an indicator of the presence of dead bacilli. Another exceptional datum we found was that a pair with negative ΔCT values in the dead-cell group was observed in our pilot study. This may have been caused by mistakes in the manual procedures such as insufficient light exposure, inappropriate shaking, or irregular, excessive pellet loss during washing. The importance of skilled sample treatment should be emphasized.
In general, smear-positive and culture-negative cases are not common. We reasoned that PMA treatment should be performed promptly because frozen storage of processed specimens damages viable cells. We therefore tested all selected smear-positive samples sequentially on the same day as we performed the requested smear and culture examination. We subsequently observed the culture results and analyzed the data. Therefore, inclusion of a sufficient number of smear-positive and culture-negative specimens is not simple. We suggest that our data can provide significant information regarding the utility of the ΔCT in detecting viable cells.
We reviewed the clinical courses of 13 patients who showed smear-positive and culture-negative results and survived to the end of our study, and calculated the time required for smear-negative conversion. A ΔCT value above 4.9 was significantly related to smear-negative conversion within 1 month from the time of the PMA real-time PCR assay. Because PMA penetrates damaged cell walls, a higher ΔCT value suggests the effectiveness of treatment regimens. Thus, it is possible to use the PMA real-time PCR to predict the treatment response. A larger scale study would be required for determining an accurate cutoff value for use in clinical mycobacteriology laboratories. In conclusion, real-time PCR with PMA is useful for differentiating dead from live bacilli in AFB smear-positive sputum samples.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2009-0066265).
References
1. Drobniewski FA, Caws M, Gibson A, Young D. Modern laboratory diagnosis of tuberculosis. Lancet Infect Dis. 2003; 3:141–147. PMID: 12614730.

2. Mishra A, Singhal A, Chauhan DS, Katoch VM, Srivastava K, Thakral SS, et al. Direct detection and identification of Mycobacterium tuberculosis and Mycobacterium bovis in bovine samples by a novel nested PCR assay: correlation with conventional techniques. J Clin Microbiol. 2005; 43:5670–5678. PMID: 16272503.
3. Su WJ. Recent advances in the molecular diagnosis of tuberculosis. J Microbiol Immunol Infect. 2002; 35:209–214. PMID: 12542245.
4. Chang HE, Heo SR, Yoo KC, Song SH, Kim SH, Kim HB, et al. Detection of Mycobacterium tuberculosis complex using real-time polymerase chain reaction. Korean J Lab Med. 2008; 28:103–108. PMID: 18458505.

5. Causse M, Ruiz P, Gutiérrez-Aroca JB, Casal M. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol. 2011; 49:3065–3067. PMID: 21653775.

6. Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med. 2003; 167:603–662. PMID: 12588714.
7. Fittipaldi M, Nocker A, Codony F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J Microbiol Methods. 2012; 91:276–289. PMID: 22940102.

8. World Health Organization. Laboratory services in tuberculosis control. Part II. Microscopy. WHO/TB/98.258. Geneva, Switzerland: World Health Organization;1998.
9. van der Kuyp F, Mahan CS. Prolonged positivity of sputum smears with negative cultures during treatment for pulmonary tuberculosis. Int J Tuberc Lung Dis. 2012; 16:1663–1667. PMID: 23131266.

10. Brescia CC, Griffin SM, Ware MW, Varughese EA, Egorov AI, Villegas EN. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl Environ Microbiol. 2009; 75:6856–6863. PMID: 19749067.
11. Yáñez MA, Nocker A, Soria-Soria E, Múrtula R, Martínez L, Catalán V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J Microbiol Methods. 2011; 85:124–130. PMID: 21329735.

12. Nocker A, Mazza A, Masson L, Camper AK, Brousseau R. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J Microbiol Methods. 2009; 76:253–261. PMID: 19103234.

13. Miotto P, Bigoni S, Migliori GB, Matteelli A, Cirillo DM. Early tuberculosis treatment monitoring by Xpert(R) MTB/RIF. Eur Respir J. 2012; 39:1269–1271. PMID: 22547737.
14. Nkuipou-Kenfack E, Engel H, Fakih S, Nocker A. Improving efficiency of viability-PCR for selective detection of live cells. J Microbiol Methods. 2013; 93:20–24. PMID: 23389080.

15. Pholwat S, Heysell S, Stroup S, Foongladda S, Houpt E. Rapid first- and second-line drug susceptibility assay for Mycobacterium tuberculosis isolates by use of quantitative PCR. J Clin Microbiol. 2011; 49:69–75. PMID: 21084506.

16. Kang H, Sung N, Lee S, Kim D, Jeon D, Hwang S, et al. Comparison of smear and culture positivity using NaOH method and NALC-NaOH method for sputum treatment. Tuberc Respir Dis. 2008; 65:379–384.

17. Pan Y. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl Environ Microbiol. 2007; 73:8028–8031. PMID: 17933922.
18. Gengenbacher M. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012; 36:514–532. PMID: 22320122.

Fig. 1
Differences between the CT values of PMA-treated and PMA-untreated aliquots of 15 Mycobacterium tuberculosis clinical isolates. Compared with the PMA-untreated live (A) group, increases in CT values in the PMA-treated live (A) group ranged from -1.5 to 4.9 and were not significant; however, compared with the results for the PMA-untreated heat inactivated (B) group, increases in the CT values in the PMA treated heat-inactivated (B) group ranged from -0.8 to 17.7 and were significant (Wilcoxon signed rank test, P=0.107, P=0.0001, respectively).
*For 1 case, a 38.9 CT value was seen in the PMA-untreated heat-inactivated (B) group, and a CT value of "not detected" was seen in the PMA-treated heat-inactivated (B) group.
Abbreviation: PMA, propidium monoazide.
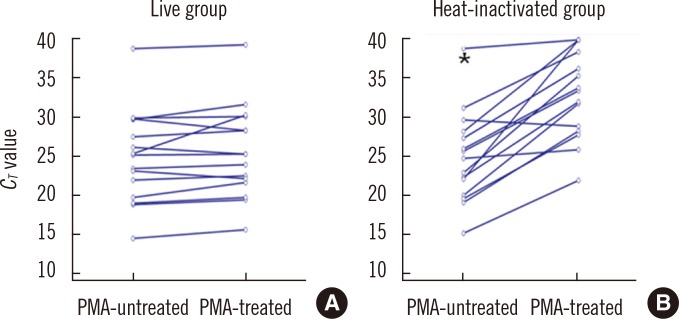
Fig. 2
ROC curve of ΔCT values in clinical respiratory specimens, in which AFB cultures were regarded as the reference method for determining the viability of cells. The area under the curve (AUC) was 0.936. The cutoff ΔCT value (*) for maximum sensitivity (89.9%) and specificity (85.7%) in determining live cells was 3.4.
Abbreviations: AFB, acid-fast bacilli; ΔCT, CT of PMA-treated specimen-CT of PMA-untreated specimen.
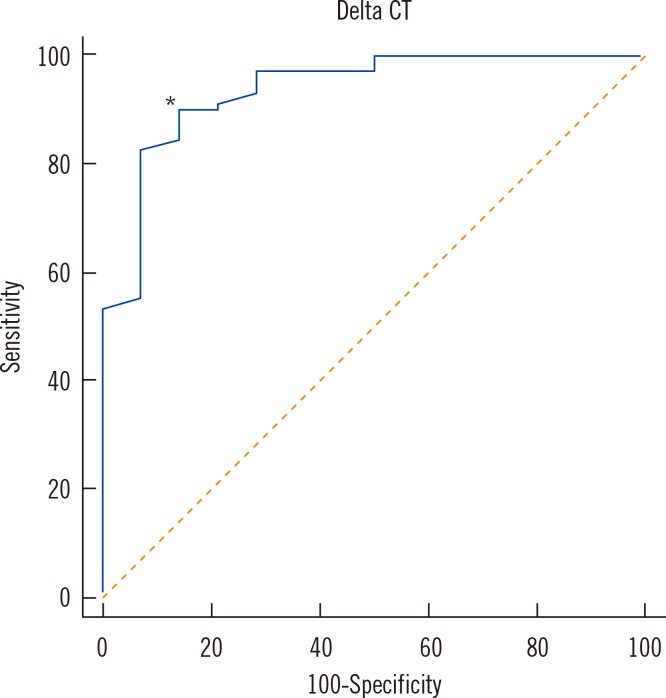
Table 1
Smear and culture results of 83 clinical respiratory specimens
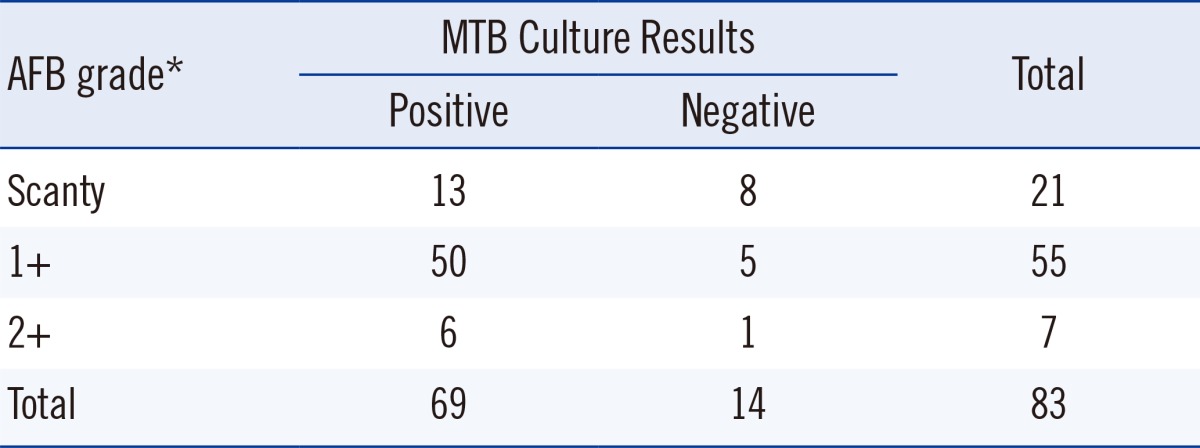
*Smear grades described according to the World Health Organization guidelines for AFB smear positivity [8].
Abbreviations: AFB, acid-fast bacilli; MTB, Mycobacterium tuberculosis.
Table 2
CT value increases after PMA treatment according to MTB culture results

*One case showed a 38.6 CT value in the PMA-untreated control specimen and a CT value of "not detected" in the PMA-treated dead-cell specimens; †The ΔCT was significantly different between the MTB culture-positive and culture-negative groups (Mann-Whitney test, P<0.0001).
Abbreviations: PMA, propidium monoazide; MTB, Mycobacterium tuberculosis; ΔCT, CT of PMA-treated specimen-CT of PMA-untreated specimen.
Table 3
Sensitivity and specificity according to various cutoff values for ΔCT in real-time PCR in PMA-treated specimens and untreated controls to predict live cells




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download