Abstract
Background
We investigated the rates of fecal transmission of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (ESBL-E) and carbapenem-resistant Enterobacteriaceae (CRE) among patients admitted to intensive care units (ICUs).
Methods
From June to August 2012, rectal cultures were acquired from all patients at ICU admission. For patients not carrying ESBL-E or CRE at admission, follow-up cultures were performed to detect acquisition. A chromogenic assay was used to screen for ESBL-E and CRE. Bacterial species identification and antibiotic susceptibility tests were performed using the Vitek 2 system (bioMérieux, France). ESBL genotypes were determined by PCR, and clonal relatedness of the isolates was assessed by pulsed-field gel electrophoresis.
Results
Out of 347 ICU admissions, 98 patients were found to be carriers of ESBL-E (28.2%, 98/347). Follow-up cultures were acquired from 91 of the patients who tested negative for ESBL-E at admission; the acquisition rate in this group was 12.1% (11/91), although none was a nosocomial transmission. For CRE, the prevalence of fecal carriage was 0.3% (1/347), and the acquisition rate was 2.9% (4/140). None of the CRE isolates were carbapenemase-producers.
Conclusions
The high prevalence of ESBL-E carriage on admission (28.2%), coupled with rare nosocomial transmission and the very low carriage rate of CRE (0.3%), challenge the routine use of active surveillance in non-epidemic settings. Nevertheless, passive surveillance measures, such as rapid and accurate screening of clinical specimens, will be critical for controlling the spread of CRE.
Infections with extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (ESBL-E) were first reported during the late 1980s [1]. Since then, the prevalence of infection with ESBL-E, notably Escherichia coli and Klebsiella pneumoniae, has increased dramatically [2, 3]. Infections caused by ESBL-E are associated with improper use of antibiotics and/or lengthy hospital stays. ESBL-E infections are also linked to increased mortality, largely due to delays in receiving effective therapy. The increasing use of carbapenems for empirical therapy of hospital-acquired sepsis has led to a rapid international dissemination of carbapenemase-producing Enterobacteriaceae (CPE), notably K. pneumoniae [4, 5].
Prior colonization is a known risk factor for ESBL-E infections [6]. The reported prevalence rate of ESBL-E varies, depending on the region and the patient population studied. In Korea, the prevalence rate of ESBL-E in clinical specimens was reported to be 12% for E. coli and 20-30% for K. pneumoniae [7, 8]. These rates are relatively higher than those reported for the Asia-Pacific region (27.7%), Latin America (23.3%), Europe (18.8%), the Middle East/Africa (16.2%), and North America (7.4%), but much lower than those reported for India (≥80%) and China (≥60%) [9, 10]. As for ESBL-E colonization in intensive care units (ICU) patients, rates varied from 2% to as high as 49% [11, 12]. Reports of carbapenem-resistant Enterobacteriaceae (CRE) have emerged during the past decade and are increasing rapidly. On the basis of the data from the Centers for Disease Control and Prevention (CDC) on bacterial species that cause healthcare-associated infections, 8% of Klebsiella species were resistant to carbapenems in 2007, a much higher figure than that seen in 2000 (<1%) [13]. In Pakistan, as many as 18.3% of patients attending a military hospital had fecal carriage of CPE; all the isolates were found to produce the New Delhi metallo-β-lactamase-1 (NDM-1) enzyme [14]. In Korea, there have also been reports of CPE, such as Citrobacter freundii and Serratia marcescens producing the VIM-2 enzyme, K. pneumoniae producing Klebsiella pneumoniae carbapenemase (KPC), and one outbreak cluster of K. pneumoniae that produced the NDM-1 enzyme [10].
In this study, we investigated the frequency of fecal carriage and acquisition rate of ESBL-E and CRE among patients admitted to ICU. In addition, we examined the molecular characteristics of the isolates. To our knowledge, this is the first study on CRE colonization rate in patients admitted to ICU in Korea.
After ethical clearance by the Institutional Review Board of the Catholic Medical Center, all patients admitted to the ICU of three teaching hospitals from June to August 2012 were included in the study. Rectal swabs were acquired at admission and thereafter at weekly intervals, or at the time of discharge. Carriers were defined as patients found to be colonized with ESBL-E or CRE at admission. To determine acquisition rates, follow-up cultures were performed for original non-carriers at weekly intervals until hospital discharge. The acquisition rate was defined as the number of patients who were not colonized at admission but became colonized afterwards, divided by the number of patients not colonized at admission.
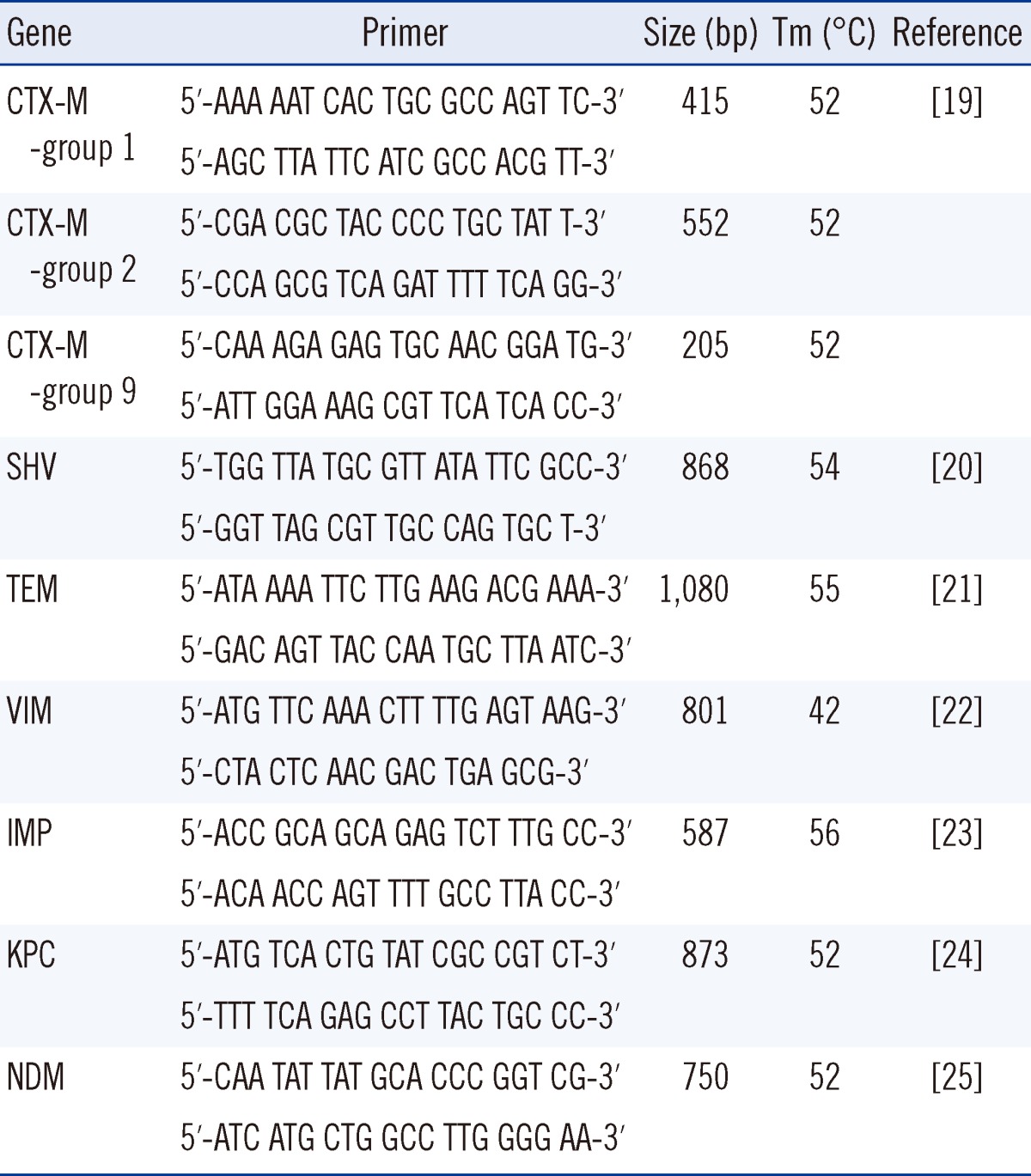
A single rectal swab specimen was collected from each patient. To detect the presence of ESBL-E, specimens were inoculated onto ChromID ESBL agar (bioMérieux, Marcy l'Etoile, France) containing 4 mg/L cefpodoxime, according to CLSI guidelines [15]. ESBL confirmatory tests were performed as described previously [16]. For CRE screening, specimens were inoculated onto KPC agar containing 1 mg/L imipenem (Rosco Diagnostica, Taastrup, Denmark) [17]. Isolates from colonies grown on the selective medium were identified to the species level and tested for antimicrobial susceptibility using the Vitek 2 system (bioMérieux). For isolates showing decreased susceptibility to imipenem or meropenem, a modified Hodge/cloverleaf test and carbapenemase confirmation test using the Rosco kit (Rosco Diagnostica) were conducted as previously described [18]. Minimum inhibitory concentrations (MICs) for ertapenem, imipenem and meropenem were determined by the broth dilution method, the E test (bioMérieux), or the disk diffusion methods, according to CLSI guidelines [15]. CRE was defined as isolates showing decreased susceptibility to ertapenem, imipenem, and/or meropenem irrespective of carbapenemase production. PCR assays were used for preliminary characterization of ESBLs present. Previously described PCR conditions and group-specific primers were used in this study (Table 1). For all isolates of a given family, clonal relatedness was determined using pulsed-field gel electrophoresis (PFGE) analysis. Briefly, after digestion with XbaI (New England Biolabs, Frankfurt, Germany), DNA fragments were separated on 1.2% agarose gels in a 0.5× TBE (Tris-borate-EDTA) buffer, using a CHEF Mapper apparatus (Bio-Rad Lab., Hercules, CA, USA). The conditions were as follows: 14℃, 6 V/cm, pulses of 10-40 sec, and a run time of 22 hr with a ramping factor of -1.357. Genetic similarities were assessed by the unweighted pair-group method using arithmetic averages (UPGMA), with the band position tolerance set at 1.8%. Tenover's criteria were used to determine whether isolates represented the same clone [26]. PFGE patterns were analyzed with the Finger-Printing II software (Bio-Rad Lab.), and were considered to be clonally related if they had a similarity coefficient greater than 80%.
When 347 patients admitted to the ICU were taken for the research, the ESBL-E carriage rate was 28.2% (98/347). Of the 249 non-carriers, follow-up rectal swabs were obtained for only 91 patients, resulting in an ESBL-E acquisition rate of 12.1% (11/91). The carriage rate of CRE was very low (0.3%, 1/347), and the acquisition rate was 2.9% (4/140). Of the 98 patients carrying ESBL-E, no patient concurrently carried CRE, although 2 patients acquired CRE afterwards.
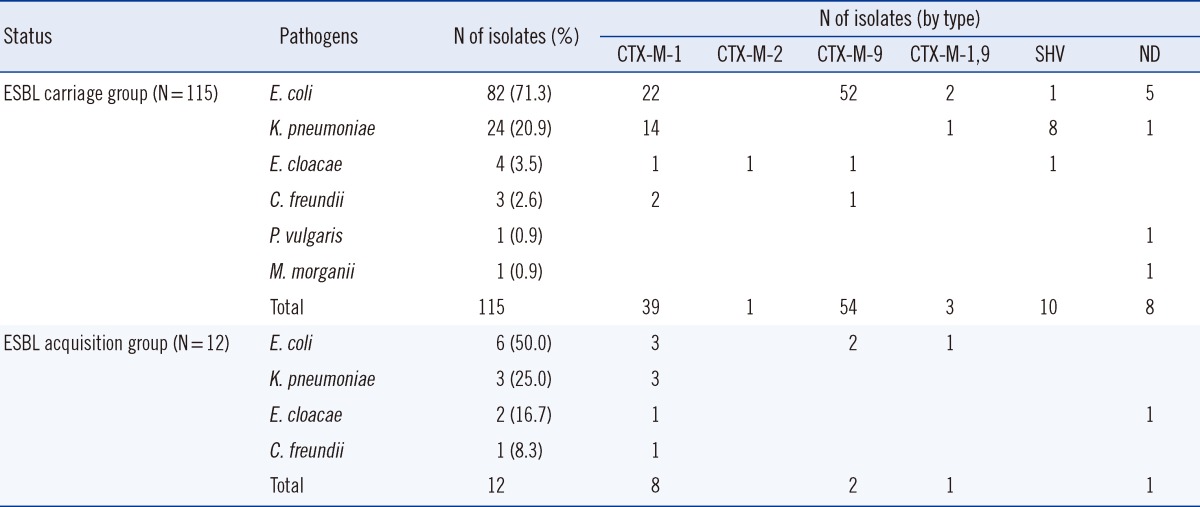
The bacterial species identified from isolates and the distribution of ESBL genotypes are shown in Table 2. E. coli and K. pneumoniae were the two most common species found among the 115 ESBL-E isolates from the carriage group (82 and 24 isolates, respectively). Of these, 76 E. coli isolates (92.7%) and 15 K. pneumoniae isolates (62.5%) harbored CTX-M type ESBLs; overall, CTX-M-9 was the most common genotype (54/115). For E. coli, CTX-M-9 was the most common type (65.9%, 54/82) whereas the CTX-M-1 type was the most common (62.5%, 15/24) for K. pneumoniae. PFGE analysis of the 82 E. coli and 24 K. pneumoniae isolates from the carriage group revealed 71 and 16 pulsotypes, respectively. Of the 12 cases who acquired ESBL-E (6 Escherichia coli, 3 K. pneumoniae, 3 Enterobacter cloacae, and 1 C. freundii), all isolates except one (E. cloacae) had CTX-M type genes. Overall, CTX-M-1 was the most common genotype (8/11). Only two CTX-M-9-producing E. coli isolates had pulsotypes closely related to isolates from the carriage group (2.2%, 2/91). However, nosocomial transmission is unlikely to account for either of these, since the carrier and acquisition cases were from different ICU, and were admitted at different times.
For CRE, only one patient carried CRE (E. coli) at admission. 4 patients acquired CRE after admission (1 Escherichia coli, 2 K. pneumoniae, and 1 Enterobacter cloacae), although none of the isolates were carbapenemase-producing.
The ESBL-E carriage rate in our study was high (28.2%, 98/347), although lower than in a recent Korean study, which reported a rate of 42.5% in ICU patients and 20.3% even in healthy persons [27]. These rates were higher than those in a 2012 report from France (15%) as well as one from Israel (8%) [28, 29]. The acquisition rate in our study was 12.1% (11/91), similar to that in the report from France (13%) and lower than that in the report from Israel (21%) [28, 29]. In the carriage group, CTX-M-9 was the most prevalent ESBL type, whereas CTX-M-1 was more prevalent in the acquisition group, particularly for E. coli isolates (E.coli-only CTX-M-1/carriage group vs. CTX-M-1/acquisition group, 22/82 vs. 3/6). There have been no reports suggesting that CTX-M-1 genes might be more effectively transferred between bacterial strains. However, given that the pandemic clone E. coli O25b:H4-ST131 has also been found to carry CTX-M-1-type genes (CTX-M-15) [30, 31], further research is needed to determine whether there are characteristics of plasmids harboring CTX-M-1-type genes that favor plasmid stability or transmission. In our study, the clonality of ESBL-E was low, and nosocomial transmission of resistant clones was not detected. Our findings are in line with a recent study involving 133 patients who were hospitalized in the same room as an ESBL-E carrier or infected person for more than 24 hr (4.3 days on average); this study found only two cases of clone transmission (1.5%) [32]. Although we identified 4-5 clones within one hospital that were not found in patient groups from other hospitals (data not shown), given that the transmission of ESBL-producing organisms can occur among family and other persons who share food or bathrooms [33], it is difficult to determine whether these clones were acquired from the local community or spread within the hospital.
Ideally, infection control for ESBL-producing strains would be achieved through active surveillance and infection control in high-risk patients, such as those in the ICU [34]. However, since both the carriage rate and the likelihood of dissemination in the local community are high, active surveillance of ESBL-E is not cost-effective, and the use of strict contact precautions for infected patients might be more appropriate.
For CRE, the carriage rate was very low (0.3%, 1/346) and the acquisition rate was 2.9% (4/140); no CPE were found in our study. However, data from the Korea Centers for Disease Control and Prevention indicate that an increasing number of hospitals are reporting the incidence of KPC- or NDM-producing strains [10, 35]. There have also been reports of horizontal transfer of KPC resistance genes between species [36, 37], notably one outbreak that involved 6 species of KPC producers (K. oxytoca, K. pneumoniae, Escherichia coli, Enterobacter cloacae, Enterobacter asburiae, and C. freundii) [37]. A recent Korean study found that a hospital where a case of NDM-1-producing K. pneumoniae was identified subsequently continued to detect the presence of NDM-1 in other bacterial species (S. marcescens and C. freundii) [38].
In conclusion, our data indicate a high fecal carriage rate for ESBL-E, but low clonality and a very low acquisition rate. Our findings suggest that in a non-epidemic setting, systematic detection of ESBL-E in ICU patients is not cost-effective, and that stringent contact precaution for infected patients might be adequate. For CRE, the fecal carriage rate was low, and no cases of CPE were found. However, given the recent emergence of CPE in several Korean hospitals and the potential for horizontal transfer of resistance genes, it will be critical to control the spread of CRE while the frequencies are still low. This will require rapid and accurate testing of carbapenem-resistant isolates from clinical specimens, as well as the adoption of appropriate infection control procedures, if carbapenemase-producing isolates are found. In addition, the incidence of CRE should be monitored continuously. If CRE incidence increases, more aggressive management, such as active surveillance of high-risk patients (those with a history of travel to high-risk countries, a history of contact with immigrants from high-risk countries, or those transferred from hospitals with an occurrence of carbapenemase-producing strains), will be necessary.
Acknowledgements
This study was supported by a grant from Korea Centers for Disease Control and Prevention in 2012 (2012 E4400200).
References
1. Kliebe C, Nies BA, Meyer JF, Tolxdorff-Neutzling RM, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985; 28:302–307. PMID: 3879659.

2. Drieux L, Brossier F, Duquesnoy O, Aubry A, Robert J, Sougakoff W, et al. Increase in hospital-acquired bloodstream infections caused by extended spectrum beta-lactamase-producing Escherichia coli in a large French teaching hospital. Eur J Clin Microbiol Infect Dis. 2009; 28:491–498. PMID: 19002728.
3. European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2010. Annual report of the European antimicrobial resistance surveillance network (EARS-Net). Updated on Nov 2011. http://ecdc.europa.eu/en/publications/Publications/1111_SUR_AMR_data.pdf.
4. Munoz-Price LS, De La Cuesta C, Adams S, Wyckoff M, Cleary T, McCurdy SP, et al. Successful eradication of a monoclonal strain of Klebsiella pneumoniae during a K. pneumoniae carbapenemase-producing K. pneumoniae outbreak in a surgical intensive care unit in Miami, Florida. Infect Control Hosp Epidemiol. 2010; 31:1074–1077. PMID: 20738186.
5. Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, et al. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol. 2011; 60:515–521. PMID: 21163830.

6. Troché G, Joly LM, Guibert M, Zazzo JF. Detection and treatment of antibiotic-resistant bacterial carriage in a surgical intensive care unit: a 6-yr prospective survey. Infect Control Hosp Epidemiol. 2005; 26:161–165. PMID: 15756887.
7. Ko KS, Lee MY, Song JH, Lee H, Jung DS, Jung SI, et al. Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated in Korean hospitals. Diagn Microbiol Infect Dis. 2008; 61:453–459. PMID: 18482815.
8. Song W, Lee H, Lee K, Jeong SH, Bae IK, Kim JS, et al. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum beta-lactamase in clinical isolates of Escherichia coli from Korea. J Med Microbiol. 2009; 58:261–266. PMID: 19141747.
9. Hoban DJ, Nicolle LE, Hawser S, Bouchillon S, Badal R. Antimicrobial susceptibility of global inpatient urinary tract isolates of Escherichia coli: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program: 2009-2010. Diagn Microbiol Infect Dis. 2011; 70:507–511. PMID: 21767706.
10. Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012; 27:128–142. PMID: 22707882.

11. Thouverez M, Talon D, Bertrand X. Control of Enterobacteriaceae producing extended-spectrum beta-lactamase in intensive care units: rectal screening may not be needed in non-epidemic situations. Infect Control Hosp Epidemiol. 2004; 25:838–841. PMID: 15518025.
12. Azim A, Dwivedi M, Rao PB, Baronia AK, Singh RK, Prasad KN, et al. Epidemiology of bacterial colonization at intensive care unit admission with emphasis on extended-spectrum beta-lactamase- and metallo-beta-lactamase-producing Gram-negative bacteria--an Indian experience. J Med Microbiol. 2010; 59:955–960. PMID: 20413621.
13. Centers for Disease Control and Prevention (CDC). Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. 2009; 58:256–260. PMID: 19300408.
14. Day KM, Ali S, Mirza IA, Sidjabat HE, Silvey A, Lanyon CV, et al. Prevalence and molecular characterization of Enterobacteriaceae producing NDM-1 carbapenemase at a military hospital in Pakistan and evaluation of two chromogenic media. Diagn Microbiol Infect Dis. 2013; 75:187–191. PMID: 23246367.
15. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-second Informational supplement, M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute;2012.
16. Song W, Jeong SH, Kim JS, Kim HS, Shin DH, Roh KH, et al. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC β-lactamases and extended-spectrum β-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn Microbiol Infect Dis. 2007; 57:315–318. PMID: 17174510.
17. Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, et al. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother. 2011; 66:2288–2294. PMID: 21788293.
18. Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007; 45:2723–2725. PMID: 17581941.
19. Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006; 57:154–155. PMID: 16284100.

20. Kim J, Kwon Y, Pai H, Kim JW, Cho DT. Survey of Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J Clin Microbiol. 1998; 36:1446–1449. PMID: 9574728.
21. Lee K, Yong D, Yum JH, Kim HH, Chong Y. Diversity of TEM-52 extended-spectrum beta-lactamase-producing non-typhoidal Salmonella isolates in Korea. J Antimicrob Chemother. 2003; 52:493–496. PMID: 12917235.
22. Poirel L, Lambert T, Türkoglü S, Ronco E, Gaillard J, Nordmann P. Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla (VIM-2) carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother. 2001; 45:546–552. PMID: 11158753.
23. Arakawa Y, Shibata N, Shibayama K, Kurokawa H, Yagi T, Fujiwara H, et al. Convenient test for screening metallo-beta-lactamase-producing gram-negative bacteria by using thiol compounds. J Clin Microbiol. 2000; 38:40–43. PMID: 10618060.
24. Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis. 2004; 39:55–60. PMID: 15206053.
25. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, bla (NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 form India. Antimicrob Agents Chemother. 2009; 53:5046–5054. PMID: 19770275.
26. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–2239. PMID: 7494007.

27. Ko YJ, Moon HW, Hur M, Park CM, Cho SE, Yun YM. Fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in Korean community and hospital settings. Infection. 2013; 41:9–13. PMID: 22723075.
28. Razazi K, DErde LP, Verachten M, Legrand P, Lesprit P, Brun-Buisson C. Clinical impact and risk factors for colonization with extended-spectrum β-lactamase-producing bacteria in the intensive care unit. Intensive Care Med. 2012; 38:1769–1778. PMID: 22893223.

29. Friedmann R, Raveh D, Zartzer E, Rudensky B, Broide E, Attias D, et al. Prospective evaluation of colonization with extended-spectrum β-lactamase (ESBL)-producing enterobacteriaceae among patients at hospital admission and of subsequent colonization with ESBL-producing enterobacteriaceae among patients during hospitalization. Infect Control Hosp Epidemiol. 2009; 30:534–542. PMID: 19419270.
30. Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, et al. Dissemination of clonality related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis. 2008; 14:195–200. PMID: 18258110.
31. Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008; 61:273–281. PMID: 18077311.
32. Tschudin-Sutter S, Frei R, Dangel M, Stranden A, Widmer AF. Rate of transmission of extended-spectrum β-lactamase-producing enterobacteriaceae without contact isolation. Clin Infect Dis. 2012; 55:1505–1511. PMID: 22955436.
33. Rodríguez-Baño J, López-Cerero L, Navarro MD, Diaz de Alba P, Pascual A. Faecal carriage of extended-spectrum β-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. J Antimicrob Chemother. 2008; 62:1142–1149. PMID: 18641033.
34. Goddard S. The efficacy of infection control interventions in reducing the incidence of extended-spectrum β-lactamase-producing Enterobacteriaceae in the nonoutbreak setting: a systematic review. Am J Infect Control. 2011; 39:599–601. PMID: 21621295.
35. Korea Centers for Disease Control & Prevention. 2011 Epidemiological report. Updated on Jun 2013. http://www.cdc.go.kr/CDC/info/CdcKrInfo0206.jsp?menuIds=HOME001-NU0004NU0026-MNU0031&fid=73&q_type=&q_value=&cid=20100&pageNum=.
36. Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010; 16:1014–1017. PMID: 20507761.
37. Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, et al. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio. 2011; 2:e00204–e00211. PMID: 22045989.

38. Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis. 2013; 76:486–490. PMID: 23688521.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download