Abstract
We aimed to observe antimicrobial resistance patterns and integron carriage of Escherichia coli isolates causing community-acquired infections. Two hundred sixty-eight E. coli strains were obtained from outpatients with various infections at different polyclinics at the 82nd Year of State Hospital in Rize, Turkey. Susceptibility to antimicrobials was tested using a disk diffusion method. The presence of integrons was examined using PCR with specific primers. Positive PCR results were confirmed by sequencing. A broth mating method was used for conjugation assays. Extragenic palindromic-PCR was performed using the oligonucleotide primer BOXA1R. Resistance frequency for ampicillin, trimethoprim/sulfamethoxazole, and tetracycline was determined as 50.6%, 33.5%, and 36.8% respectively. No strains were resistant to amikacin. Seventy isolates were positive for the intI1 gene, of which 49 carried gene cassettes. Eleven isolates were positive for the intI2 gene, eight of which carried gene cassettes. Seven gene cassettes (dfrA1, dfrA5, dfrA7, dfrA17, aadA1, aadA5, and sat2) were predominantly harbored in integrons. We detected conjugative plasmids harboring integrons in two E. coli strains. Four strain clusters were yielded by BOX-PCR fingerprints showing that they were clonally related. No apparent relationship occurred among class 1 and 2 integron-carrying strains. We conclude that integrons are widespread in genetically variable E. coli strains and will continue to mediate dissemination of resistance genes in the community.
Escherichia coli is one of the most important human pathogens, responsible for up to 90% of all community-acquired urinary tract infections (UTIs) [1]. The emergence and spread of antimicrobial-resistant E. coli has become a serious public health threat worldwide [2]. Integrons are among the main types of mobile elements collectively known as a natural gene capture system in bacteria. Class 1 integrons consist of both a 5'-conserved segment carrying the gene intI encoding a site-specific integrase protein and a variable region containing one or more antibiotic resistance genes. Class 2 integrons are related to a mobile genetic element, Tn7 transposon, and their structures resemble those of class 1 integrons [3]. Several studies have been focused on investigating the carriage of integrons and the resistance frequency of E. coli strains causing community-acquired infections [4]. The goal of the present study was to screen the antibiotic resistance genes inserted into class 1 and 2 integrons in E. coli isolates obtained from clinical samples.
Two hundred sixty-eight antibiotic-resistant E. coli strains were isolated from clinical specimens (mainly urine) of unrelated outpatients with different infections at various policlinics at the 82nd Year of State Hospital in Rize, Turkey, between March 2011 and February 2012. Bacterial identification was carried out using both conventional methods and Vitek2 compact system (bioMerieux, Durham, NC, USA) according to the manufacturers' instructions. Susceptibility to antimicrobials was tested with a disk diffusion method according to the Clinical and Laboratory Standards Institute guideline [5]. The following antimicrobials (Oxoid, Basingstoke, UK) were used: amikacin (30 µg), ampicillin (10 µg), ampicillin/sulbactam (20 µg), aztreonam (30 µg), ceftazidime (30 µg), ceftriaxone (30 µg), cefuroxime (30 µg), ciprofloxacin (5 µg), imipenem (10 µg), norfloxacin (10 µg), ofloxacin (5 µg), streptomycin (10 µg), gentamicin (30 µg), tetracycline (10 µg), and trimethoprim/sulfamethoxazole (1.25 µg/23.75 µg).
DNA templates for PCR were prepared by boiling the bacterial cultures. The presence of integrons was examined using PCR with specific primers. Reaction composition and cycling parameters were as previously described [6, 7]. After the PCR amplicons were purified with purification kit (Qiagen, Courtaboeuf, France), they were cloned and sent to Macrogen (Seoul, Korea) for DNA sequencing. Sequencing data were evaluated using the Basic Local Alignment Search Tool from the National Center for Biotechnology Information website [8]. Conjugative plasmids harboring integrons were screened with conjugation assays using a broth mating method [9]. Recipient to transconjugant ratio gave to the conjugation efficiency. Repetitive extragenic palindromic-PCR was performed using the oligonucleotide primer BOXA1R (BOX-PCR) [10]. Reaction conditions and optimizations were performed as described elsewhere [11]. BOX fingerprints were analyzed using a Phoretix gel analysis package (Nonlinear USA Inc., Durham, NC, USA) to show similarity among the strains.
We evaluated 268 nonrepetitive (one per patient) E. coli isolates in this study, and the total resistance frequency of the isolates is shown in Fig. 1. Resistance to ampicillin was prevalent in 50.6% of the isolates. Resistance to streptomycin (27.9%), trimethoprim/sulfamethoxazole (33.5%), and tetracycline (36.8%) were also high. The lowest resistance rate was to aztreonam (7.4%), and resistance to amikacin was not observed.
Of 268 strains, 81 (30.2%) were positive for intI1 and intI2. The variable regions of the integrons were amplified for 49 (18.28%) of 70 intI1-positive strains and eight (2.98%) of 11 intI2-positive strains (Table 1). The amplicon length of the integrons varied between 700 and 2,200 bp. Seven different gene cassettes, including dfrA1, dfrA5, dfrA7, dfrA17, aadA1, aadA5, and sat2, were observed to contain dfr, aad, and sat gene alleles conferring resistance to trimethoprim and aminoglycosides. The most prevalent gene cassette array found, dfrA17-aadA5, accounted for approximately 47% of the sequenced cassettes (Table 1). Two class 1 integrons were found on two separate conjugative plasmids in strains 76 and 226 from outpatients at the pediatrics and urology clinics, respectively. Their conjugal transfer frequency ranged from 10-6 to 10-2 (data not shown). These two strains are clonally unrelated and co-transferred resistance to ampicillin, ceftriaxone, trimethoprim/sulfamethoxazole, gentamicin, and tetracycline (Table 1). BOX-PCR genotyping analyses showed that PCR products with various molecular weights obtained from the strains were discriminated from one another, and the 49 strains were phylogenetically clustered into four main clusters (A to D) (Fig. 2). Of 47 strains, 30 were clustered in group A, 12 in group B, 5 in group C., and two in group D. All class 2 integron-carrying E. coli strains (n=7) were collected in cluster A.
Typing by DNA fingerprinting is a common tool for epidemiology studies and bacterial species or isolates are distinguished by BOX-PCR pattern. BOX-PCR is among the most used techniques in biogeography studies of microbial isolates. The data obtained from DNA amplification from E. coli strains allowed the distinction of four main BOX clusters according to genotype differences. It may be expected that all class 2 carrying-E. coli strains would be clustered in a separate branch. In the present study, all class 2 carrying-E. coli strains were clustered with the most of the class 1 carrying-E. coli strains (n=23) and BOX-PCR could separate class 2 carrying-E. coli strains from some of the class 1 carrying-E. coli strains. The clustering of strains was not due to dissemination of specific clones but due to the possible acquisition of genes through unrelated clones. An earlier study performed in this state hospital detected 12 pulsed-field gel electrophoresis types in class 1 integron-carrying E. coli strains, and the 11 type patterns were indistinguishable [4]. A linkage between resistance genes and others that contribute to increased bacterial survival may also aid the maintenance of resistance genes in the same strain. Other epidemiological characteristics, such as specimens isolated, isolation date, and unit of the integron-containing strains are included in Table 1.
We found resistance to various groups of antimicrobials, which occurs most often among clinical isolates from nosocomial infections. On the contrary, all of the clinical isolates were cultured from outpatients. It could be assumed that resistance to such antimicrobials may be partially due to selection imposed by selective antibiotic pressure on populations. More than a half of the strains were isolated from the urine samples of pediatric patients with UTIs. The results of antimicrobial susceptibility test are in general agreement with those reported by El-Najjar et al. [12], who found the highest levels of resistance to ampicillin (60%), trimethoprim/sulfamethoxazole (55%), streptomycin (53%), and tetracycline (51%). Sawma-Awad et al. [13] also reported high levels of resistance in a Lebanese population to ampicillin (68%), tetracycline (43%), and trimethoprim/sulfamethoxazole (38%) and resistance levels for ciprofloxacin (16%) and gentamicin (12%) similar to those obtained in this study.
Although several studies have addressed the emergence of integron-harboring Enterobacteriaceae of environmental origin in Turkey [14, 15], only limited epidemiological surveys have been carried out until now [4]. The current study revealed a frequency (30.2%) that was quite high for intI among clinical E. coli strains isolated mainly from UTIs. Some studies have reported that class 1 and 2 integrons are widespread in uropathogenic E. coli isolates from urinary samples of outpatients [16]. The percentage of the antibiotic-resistant strains is comparable to those of other studies.that is, 43-75% of the resistant clinical bacteria with integrons [7, 17]. Horizontal dissemination of resistance genes has been concluded to occur very efficiently among members of family Enterobacteriaceae in nosocomial settings [18]. By comparison, we detected a very low rate of conjugative plasmids harboring integrons in E. coli strains of community-acquired infections.
Most of these strains also harbored class 1 and 2 integrons, and seventy intI-positive strains showed multiple antibiotic resistance phenotypes, whereas a majority of the other 187 strains without integrons did not (data not shown). We believe that the present findings will augment understanding of the factors contributing to the existence and dissemination of antimicrobial resistance encoded by the integrons in E. coli UTIs, although we also detected multiple antibiotic resistances in strains with empty integrons, such as those containing the integrons with gene cassettes. Studying integrons and their resistance phenotypes can provide important information on the mechanisms of acquisition of multiple antibiotic resistance genes in clinical isolates of community-acquired infections.
Acknowledgments
This work was supported by Recep Tayyip Erdogan University Research Fund grants BAP-2012.106.01.11 and BAP-2011.102.03.3.
References
1. Jones RN, Kugler KC, Pfaller MA, Winokur PL. Characteristics of pathogens causing urinary tract infections in hospitals in North America: results from the SENTRY Antimicrobial Surveillance Program, 1997. Diagn Microbiol Infect Dis. 1999; 35:55–63. PMID: 10529882.

2. Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol. 2009; 12:476–481. PMID: 19716760.

3. Cocchi S, Grasselli E, Gutacker M, Benagli C, Convert M, Piffaretti JC. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol. 2007; 50:126–132. PMID: 17456180.
4. Sandalli C, Buruk CK, Sancaktar M, Ozgumus OB. Prevalence of integrons and a new dfrA17 variant in Gram-negative bacilli that cause community-acquired infections. Microbiol Immunol. 2010; 54:164–169. PMID: 20236427.
5. National Committee for Clinical Laboratory Standards. Tenth Informational supplement, M100-S10 (M2). Performance standards for antimicrobial susceptibility testing. Wayne, PA: National Committee for Clinical Laboratory Standards;2000.
6. Lévesque C, Piché L, Larose C, Roy PH. PCR Mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995; 39:185–191. PMID: 7695304.

7. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001; 45:2658–2661. PMID: 11502548.
9. Rice LB, Willey SH, Papanicolaou GA, Medeiros AA, Eliopoulos GM, Moellering RC Jr, et al. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemother. 1990; 34:2193–2199. PMID: 2073110.
10. Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994; 5:25–40.
11. Seurinck S, Verstraete W, Siciliano SD. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl Environ Microbiol. 2003; 69:4942–4950. PMID: 12902290.
12. El-Najjar NG, Farah MJ, Hashwa FA, Tokajian ST. Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. Lett Appl Microbiol. 2010; 51:456–461. PMID: 20840552.
13. Sawma-Aouad G, Hashwa F, Tokajian S. Antimicrobial resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli in Lebanon. J Chemother. 2009; 21:153–158. PMID: 19423467.
14. Ozgumus OB, Celik-Sevim E, Alpay-Karaoglu S, Sandalli C, Sevim A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J Microbiol. 2007; 45:379–387. PMID: 17978796.
15. Ozgumus OB, Sandalli C, Sevim A, Celik-Sevim E, Sivri N. Class 1 and class 2 integrons and plasmid-mediated antibiotic resistance in coliforms isolated from ten rivers in northern Turkey. J Microbiol. 2009; 47:19–27. PMID: 19229487.

16. Solberg OD, Ajiboye RM, Riley LW. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J Clin Microbiol. 2006; 44:1347–1351. PMID: 16597861.
17. Jones LA, McIver CJ, Rawlinson WD, White PA. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intell Q Rep. 2003; 27(Suppl):S103–S110. PMID: 12807284.
18. Leverstein-van Hall MA, Box AT, Blok HE, Paauw A, Fluit AC, Verhoef J. Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J Infect Dis. 2002; 186:49–56. PMID: 12089661.

Fig. 1
Total antimicrobial resistance frequencies of 268 clinical isolates of Escherichia coli.
Abbreviations: AK, amikacin; AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CN, gentamicin; CRO, ceftriaxone; CXM, cefuroxime; IPM, imipenem; NOR, norfloxacin; OFX, ofloxacin; S, streptomycin; SAM, ampicillin/sulbactam; SXT, trimethoprim/sulfamethoxazole; TE, tetracycline.
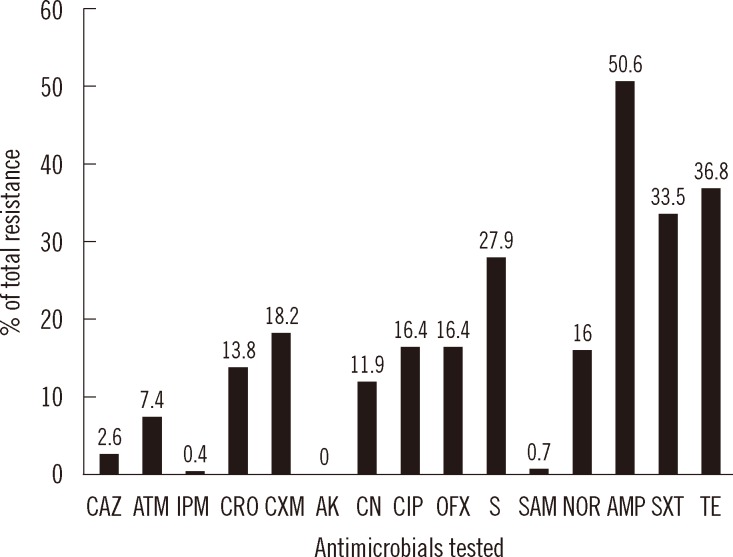
Fig. 2
Dendrogram analysis of E. coli strains harboring class 1 and 2 integrons using repetitive extragenic palindromic-PCR with the oligonucleotide primer BOXA1R. All strains clustered into four groups (A to D), as seen in Table 1. While 42 class 1 integron-carrying E. coli strains were collected in four different clusters, class 2 integron-carrying E. coli strains (13, 48, 172, 185, 283, 305, and 684) were collected only in the cluster A.
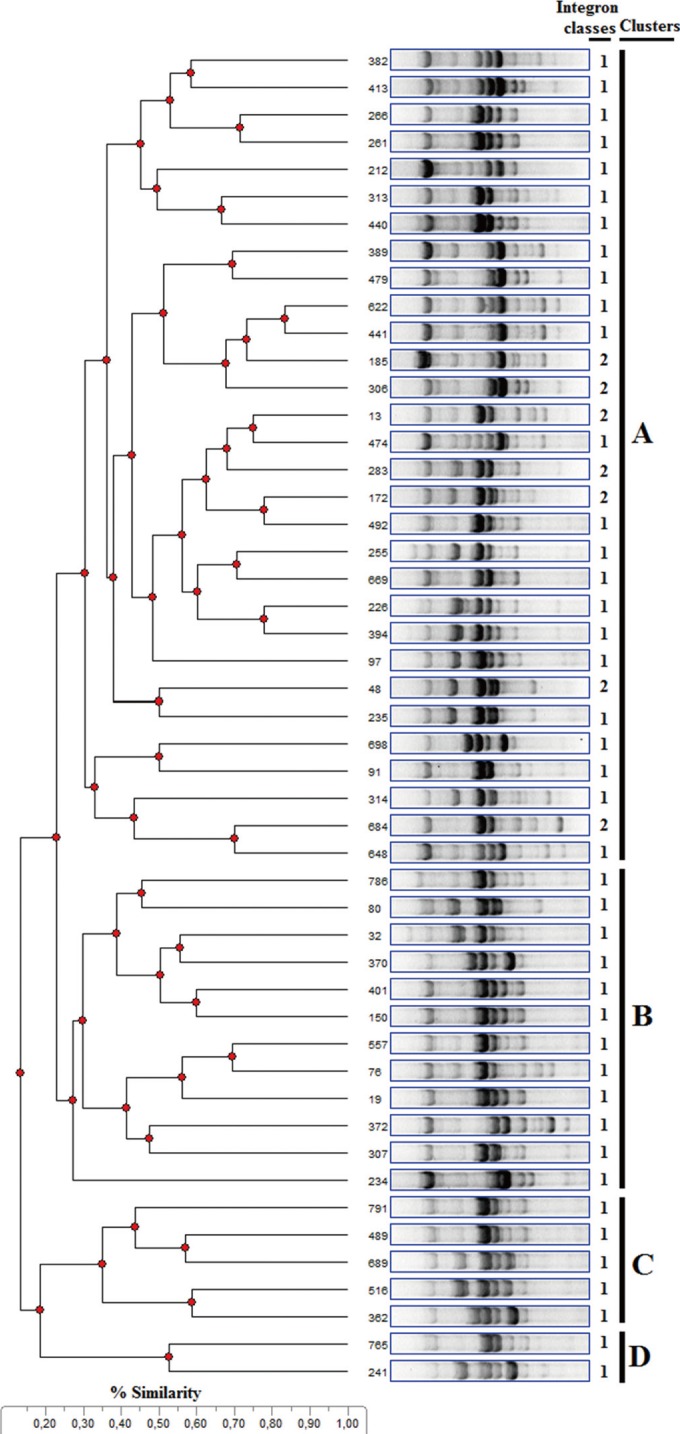
Table 1
Epidemiological characteristics of 49 Escherichia coli strains causing community-acquired infections harboring class 1 and 2 integrons
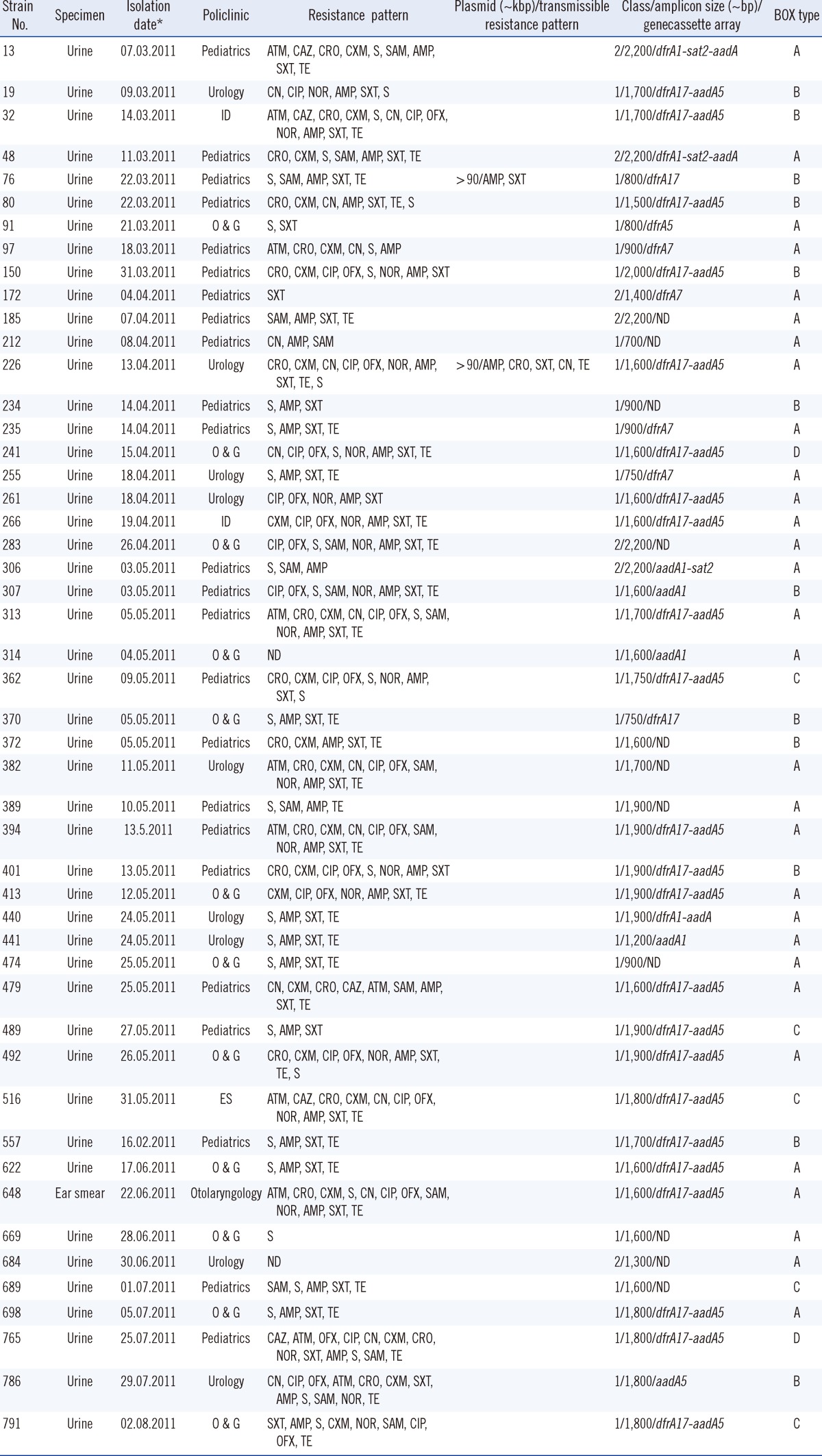
*Day. month. year.
Abbreviations: AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CN, gentamicin; CRO, ceftriaxone; CXM, cefuroxime; ES, emergency service; ID, infectious disease; ND, not detected; NOR, norfloxacin; O & G, obstetrics and gynecology; OFX, ofloxacin; S, streptomycin; SAM, sulbactam/ampicillin; SXT, trimethoprim/sulfamethoxazole; TE, tetracycline.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download