Abstract
Background
Clinical specificity and sensitivity are essential factors in the adoption of a human papillomavirus (HPV) test as a primary screening tool and test of cure after treatment of cervical cancer and precancerous lesions (High-Risk-Lesion). Using histologically-confirmed High-Risk-Lesion-patient specimens with postoperative follow-ups, we performed clinical validation of the AdvanSure GenoBlot Assay (GenoBlot; LG Life Sciences, Korea).
Methods
The study population included 100 cases with High-Risk-Lesion, 96 with high-risk genotype positive and cervical intraepithelial neoplasia (CIN) 1 or better, and 39 with HR-negative and better than CIN 1. Forty-eight High-Risk-Lesion cases received follow-up HPV exams after surgery. For validation as a test of cure, 48 preoperative specimens (PreOP) and 78 postoperative specimens (PostOP) from 48 subjects were separately analyzed. The results of HPV DNA chip tests (HPVDNAChip; BioMedLab Co., Korea) and sequencing were cross-compared.
Results
The concordance rates for each genotype between HPVDNAChip and GenoBlot were between 96.3-100%. The accuracy of HPVDNAChip and GenoBlot was 87.9% and 96.6%, respectively. Genotype-based specificity for High-Risk-Lesion detection was higher than 87% for both assays; genotype 16 showed the highest sensitivity. In the PostOP group, the positive rates for HPVDNAChip and GenoBlot were 30.8% and 47.4%, respectively.
The factors that determine the adoption of human papillomavirus (HPV) test as a screening tool include its accompanying clinical sensitivity and specificity, as well as its analytical sensitivity [1]. In current practice, there are three major contexts in which tests for high risk (HR) genotypes of HPV are relevant: 1) primary screening of cervical cancer and precancerous lesions; 2) triage of low-grade disease; and 3) test of cure after treatment [2, 3]. A significant number of studies have been reported for the primary cervical high risk lesion screen and triage; however, little has been reported in terms of the evaluation of test of cure [2-11].
While HPV HR have been continually updated, notorious HR such as 16 and 18 are stable. Other genotypes, such as 26, 53, 61, 62, 69, 70, 73, and 82, are alternately classified as either high or low-risk groups [12-14]. AdvanSure GenoBlot Assay (GenoBlot; LG Life Sciences, Seoul, Korea) is a system of real-time PCR and reverse hybridization detecting 20 HR and 15 low-risk HPV types. It detects comparatively more HR HPV types-including 26, 53, 69, 70, 73, and 82 that are gradually being classified as HR types [13, 14]-than genotyping systems currently available on the market; however, it has not yet been clinically validated.
In this study, we used a population that contains patients with low-grade lesions and patients with histologically-confirmed, high risk lesions in order to evaluate GenoBlot as a primary screening tool. The specimens from high risk lesion population include not only the samples obtained at diagnosis but also those acquired after surgical interventions, to evaluate the validity of GenoBlot as a test of cure. The results of HPV DNA chip tests (HPVDNAChip; BioMedLab Co., Seoul, Korea) and sequencing were cross-compared for analytical validation.
The study subjects were 235 patients with a mean age of 44 yr and an age range of 16-78 who underwent histological exams and HPVDNAChip analysis at the Catholic University Bucheon St. Mary's Hospital between January 2011 and November 2012. Among the subjects, there were 100 cases with cervical intraepithelial neoplasia (CIN) 2 or worse in biopsy (High-Risk-Lesion group), 96 with HR-positive and CIN 1 or better (HR-Positive Benign group), and 39 with HR-negative and better than CIN 1 (HR-Negative Benign group).
The total number of specimens was 379, 187 specimens from the High-Risk-Lesion group (47 cases with one specimen each and 53 cases with 140 follow-up specimens), 130 specimens from the HR-Positive Benign group (68 cases with one specimen each and 28 cases with 62 follow-up specimens), and 62 specimens from the HR-Negative Benign group (21 cases with one specimen each and 18 cases with 41 follow-up specimens).
Among the High-Risk-Lesion subjects, 95 cases underwent loop electrosurgical excision procedure (LEEP) conization of the cervix or total hysterectomy, and 48 cases received HPV exams more than once after the surgical operations. For validation as a test of cure, these 48 preoperative specimens (PreOP) and 78 postoperative specimens (PostOP) from 48 subjects were separately analyzed. The 48 subjects received an operation within 15 days after the initial diagnosis and were subjected to postoperative follow-up with an interval of 3-5 months. The total duration of follow-up was in the range of 6-32 months, with a median duration of 16.3 months.
This study was fully approved by the Catholic Medical Center institutional review board.
Using a sterilized vaginal speculum, cervical specimens were collected by inserting a cytobrush attached to an HPVDNAChip Sampler (BioMedLab Co.) into the endo-, and exo-cervical areas. After removal, the cytobrush was placed into transport medium (1× phosphate buffered saline) and immediately refrigerated. DNAs extracted in compliance with the manufacturer's instructions was subjected to immediate use or stored at 4℃, and in case of longer than one day of storage, stored at -70℃.
All HPV assays were conducted by using a PCR-based DNA microarray system, HPVDNAChip that provides 22 HPV probes: 15 HR (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 69) and 7 low-risk (6, 11, 34, 40, 42, 43, and 44) types. The assays followed the manufacturer's protocol [15]. A DNA chip scanner (GenePix Pro 3.10; GenePix 4000B, Laser power-33%, PMT-880; Axon Instrument Inc., Union City, CA, USA) was used to visualize the hybridized HPV DNA.
GenoBlot is a system for the detection and discrimination of 20 HR (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 70, 73, 82) types, and 15 low-risk (6, 11, 32, 34, 40, 42, 43, 44, 54, 55, 57, 61, 62, 81, 83) types in each strip with a combination of real-time PCR and reverse hybridization technologies.
Single-tube nested asymmetric real-time PCR was performed according to the manufacturer's protocol. For the real-time PCR-positive samples, hybridization and interpretation were performed by Genoline Station (LG Life Sciences).
Sequencing was performed when HPVDNAChip and GenoBlot showed different results for a sample and when GenoBlot showed positive results for a newly-included genotype. All PostOP samples were also subjected to sequencing. Two types of primers (MY and gp) were designed by using internal fragments of the HPV L1 region. PCR was performed by using 13 µL of HS Taq premix (GenetBio, Daejeon, Korea), 2 µL of each primer set (10 µM) and 5 µL of extracted template DNA in a PTC-200 (BIO-RAD, Hercules, CA, USA) under the following conditions: 95℃ for 10 min followed by 35 cycles of 95℃ for 15 sec, 55℃ for 45 sec, and 72℃ for 30 sec for the MY region; or 95℃ for 10 min followed by 35 cycles of 95℃ for 15 sec, 48℃ for 45 sec, and 72℃ for 30 sec for the gp region (nested PCR). A final extension step was performed for each PCR at 72℃ for 5 min. The PCR products were purified by using QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sequencing of positive PCR samples was performed by ABI Prism 3700 (Applied Biosystems, Foster City, CA, USA) and the DNA sequences were analyzed by basic local alignment search tool (BLAST).
SAS System for Windows v8.02 was used to run McNemar's test to evaluate accuracy based on the true value of sequencing. Concordance rates, kappa values, prevalence adjusted bias adjusted kappa (PABAK), specificity, and sensitivity were calculated based on the results of biopsy, HPVDNAChip, and sequencing. The confidence interval (CI) was set to 95% and P value less than 0.05.
Table 1 shows the positive rate of each genotype by group and for the 379 specimens.
The genotypes that showed higher positive rates in the High-Risk-Lesion group than in the HR-positive Benign group are 16 and 33 by HPVDNAChip, and 16, 18, and 33 by GenoBlot. The genotypes of 51, 56, and 66 showed lower positive rates in the High-Risk-Lesion group than in the HR-positive Benign group for both assays.
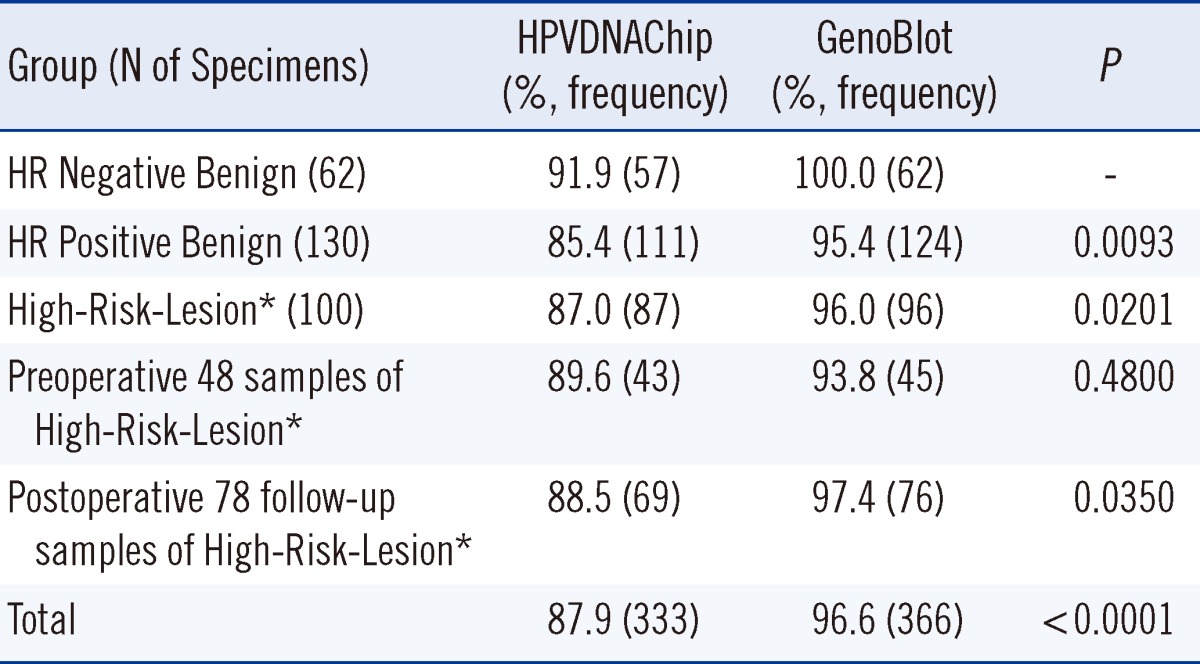
Table 2 compares the concordance rates, kappa values and PABAKs of HPVDNAChip and GenoBlot in each group and for the 379 specimens. The concordance rate for each genotype fell in the range of 96.3-100%, and the High-Risk-Lesion group showed concordance rates of 93.0-100%.
Accuracy, in contrast to the sequencing results, of HPVDNAChip and GenoBlot is shown in Table 3. The accuracy is generally higher across all groups analyzed by GenoBlot than in those analyzed by HPVDNAChip: 87.9% accuracy by HPVDNAChip and 96.6% by GenoBlot.
Table 4 shows the specificity and sensitivity of HPVDNAChip and GenoBlot in light of High-Risk-Lesion diagnosis. Genotype-based specificity was higher than 87% for both assays: genotypes 16, 33, 35, 39, 52, 58, and 66 showed higher specificity by HPVDNAChip analysis than by GenoBlot, with the exception that GenoBlot exhibited higher specificity for genotypes 51 and 56. Genotype 16 demonstrated the highest sensitivity among all the genotypes.
Forty-eight subjects, including PostOP follow-up specimens, were separately analyzed in two groups: 48 PreOP specimens were collected from the High-Risk-Lesion, and 78 PostOP follow-up specimens were collected. The number of subjects with positive result in the first follow-up HPV tests after their operation was 15 by HPVDNAChip and 22 by GenoBlot, 9 cases of which had infections with new genotypes. There were 9 cases that were positive until 9 months after their operation (up to 3 follow-up tests), 4 of which had infections with new genotypes and 5 of which had either infections with persistent genotypes or multiple infections involving persistent genotypes. Among the 48 cases, 3 cases received a second operation for relapse after previously testing positive in their first follow-up HPV tests after operation and being infected with the same genotypes. One of the 3 cases expired by metastasis.
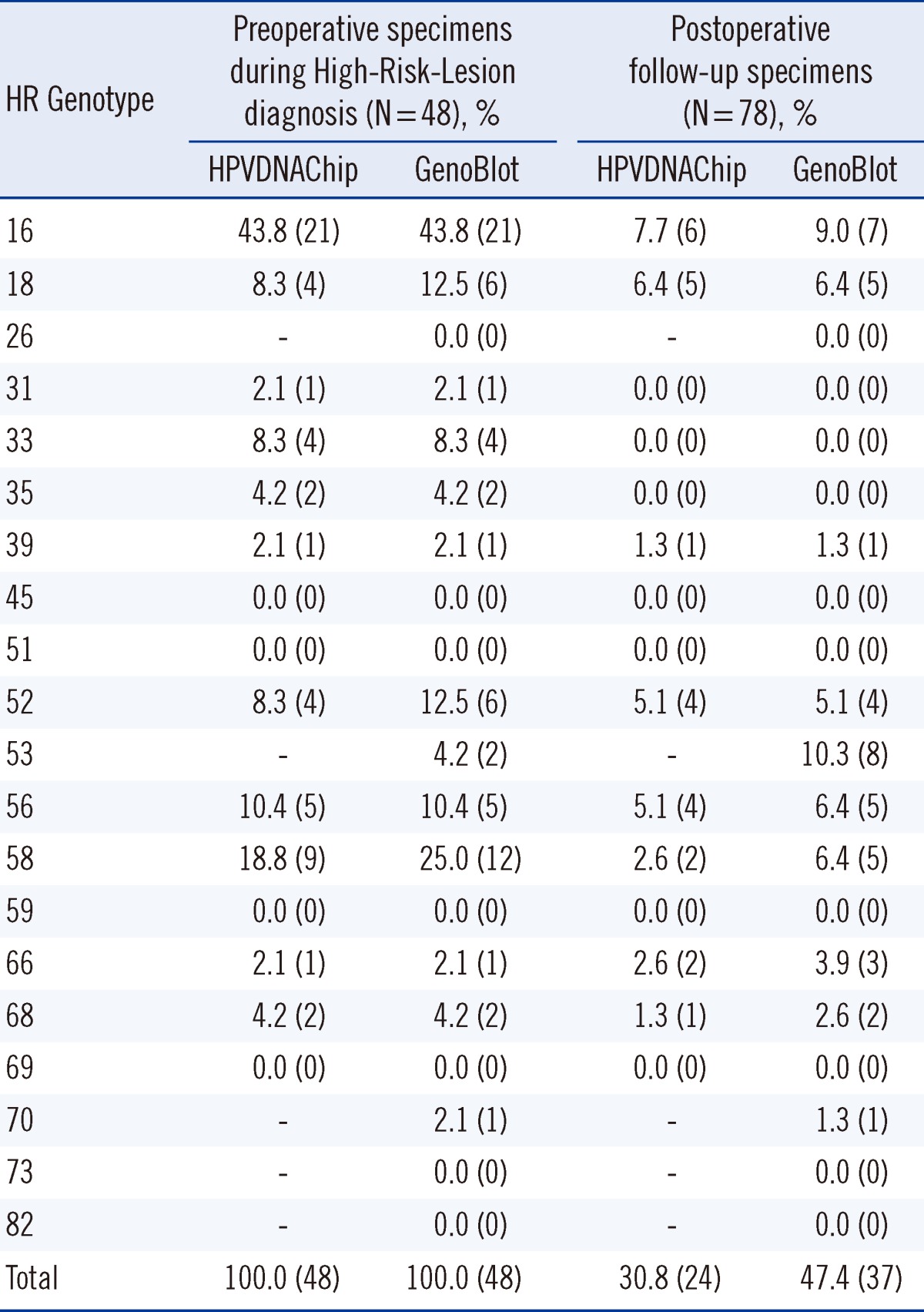
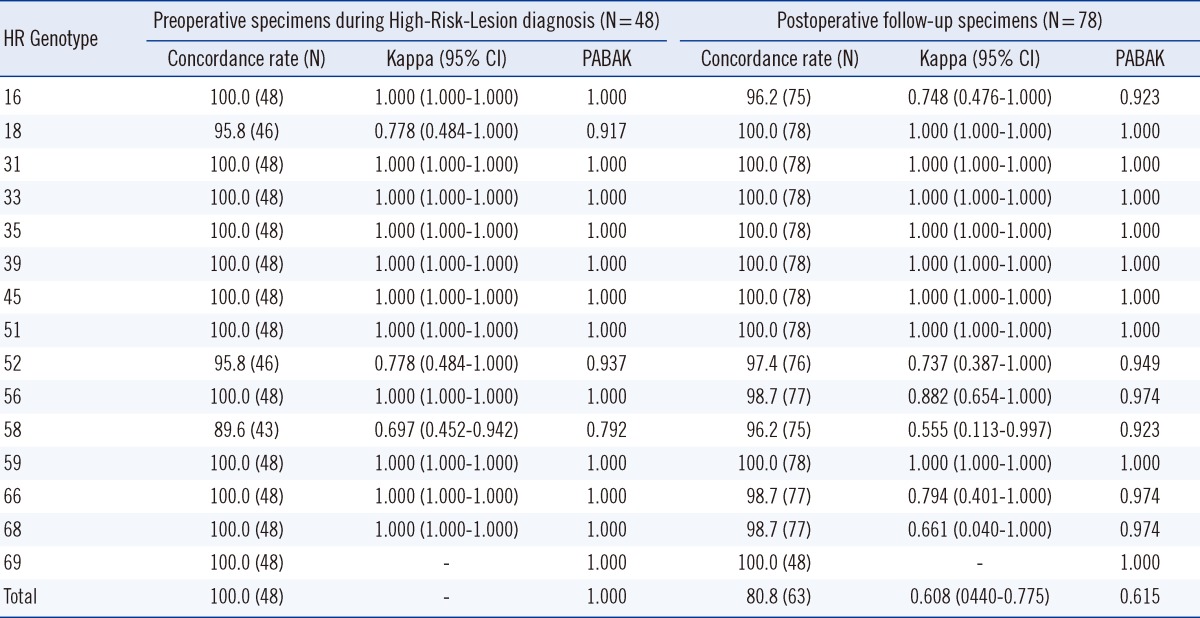
Tables 5 and 6 show the positive and concordance rates of each genotype in the groups. In the PostOP group, the positive rates for HPVDNAChip and GenoBlot were 30.8% and 47.4%, respectively, with GenoBlot showing a significantly higher rate (P=0.0348). The accuracy of both assays compared with sequencing is shown in Table 3. Compared to the similar accuracy of both assays in the PreOP specimens, GenoBlot showed a higher accuracy than HPVDNAChip in the PostOP specimens.
The overall positive rates and the concordance rate of the two assays were comparable to those of previous reports [4-11]. The positive rates of individual genotypes were relatively higher in GenoBlot than in HPVDNAChip, with exceptions for genotypes 51 and 56, for which HPVDNAChip showed higher positive rates than GenoBlot. Hence, when HR types were arranged in terms of frequency, HPVDNAChip showed 16 as the most frequent genotype followed by 58, 52, 51, 18, and 56, whereas GenoBlot listed 16, 58, 52, 66, 18, and 35.
Among the 16 samples that were determined to be positive only by GenoBlot, 8 were of type 53 (only available in GenoBlot), 1 of type 70, 1 of type 16, 3 of type 58, 1 of type 56, 1 of type 66, and 1 of type 68.
There were 2 cases of type 16 that resulted in discrepancy: the HPVDNAChip tests were negative, whereas the GenoBlot as well as sequencing, were positive in both cases. In 1 type 18 case, the HPVDNAChip test was negative, while both the GenoBlot and sequencing were positive, indicating that the HPVDNAChip failed to detect the critical HR types 16 and 18, especially in 2 cases of High-Risk-Lesion, necessitating an increase in its detection sensitivity.
Among the 100 High-Risk-Lesion cases, 3 cases were negative in HPVDNAChip, all of which were positive in GenoBlot: one case of single infection with type 16, one case of single infection with type 53, and one case of multiple infections with types 16, 56, and 70.
The specificity of GenoBlot and HPVDNAChip was sufficient as a screening tool and similar to the previous findings [4, 9, 16].
Among the GenoBlot's newly-included HR types, 26 and 73 were not detected from the samples included in the study but 23 cases of type 53, 11 cases of type 70, and 2 cases of type 82 were detected. Of the type 53-positive cases, one case, which was negative in the HPVDNAChip test from the sample at the time of High-Risk-Lesion diagnosis, was shown to be infected with the single genotype. This demonstrates that it is worth considering additional HR types when predicting High-Risk-Lesions before biopsy or as an infomration resource that promotes patient compliance with future follow-up exams.
The difficulty in following up the patients with LEEP conization contributed to the loss of cervical os boundary areas, which makes detecting re-infection, relapse, or persistence of the lesion a challenge. Negative conversion in HPV tests after surgical treatments reflects the appropriateness of the clinical sensitivity of HPV test results. Clinical implications vary among such cases as when negative results are obtained immediately after surgical treatments such as conization, when remaining DNA is cleared over a time to negative conversion, and in the persistence of infection or re-infection.
In this study, the 100 High-Risk-Lesion cases included 48 cases, in which the HPV test results were secured during High-Risk-Lesion diagnosis or LEEP conization or after total hysterectomy. From these 48 cases, when the follow-up samples after surgery were examined, 15 cases (31.3%) showed HPV-positive results by HPVDNAChip and 22 cases (45.8%) by GenoBlot. Among the 78 follow-up samples, 24 samples (30.8%) were HPV positive by HPVDNAChip and 37 samples (47.4%) by GenoBlot. In these post-surgery HPV-positive cases, re-infection was confirmed in 2 cases due to, for example, deteriorating lesion, and for which total hysterectomy was performed. There were, due to infection by new genotypes, 9 cases with re-infection; 3 were confirmed by HPVDNAChip and 6 by GenoBlot.
This result strongly establishes the fact that the follow-up exams of patients are of definitive help in determining whether previous genotypes persist or new types bring about new infections. However, further analysis would be required to determine the appropriate levels of clinical sensitivity in detecting small amounts of remaining DNA.
In conclusion, GenoBlot is capable of detecting 35 gentotypes sufficiently and demonstrates valid clinical specificities higher than HPVDNAChip; GenoBlot can potentially replace HPVDNAChip tests for 22 genotypes. The newly-included genotypes were proven to assist in predicting High-Risk-Lesion, and as a test of cure, GenoBlot was more sensitive than the HPVDNAChip.
References
1. Arbyn M, Ronco G, Meijer CJ, Naucler P. Trails comparing cytology with human papillomavirus screening. Lancet Oncol. 2009; 10:935–936. PMID: 19796748.
2. Poljak M, Oštrbenk A. The Abbott RealTime High Risk HPV test is a clinically validated human papillomavirus assay for triage in the referral population and use in primary cervical cancer screening in women 30 years and older: a review of validation studies. Acta Dermatovenerol Alp Panonica Adriat. 2013; 22:43–47. PMID: 23836358.
3. Cuschieri K, Hardie A, Hovland S, Hoaas B, Karlsen F, Cubie H. Comparison of the sensitivities of three commercial assays for detection of the high risk HPV types 16, 18 and 45. J Virol Methods. 2013; 193:147–150. PMID: 23727117.

4. Lloveras B, Gomez S, Alameda F, Bellosillo B, Mojal S, Muset M, et al. HPV testing by cobas HPV test in a population from Catalonia. PLoS One. 2013; 8:e58153. PMID: 23483984.

5. Castle PE, Stoler MH, Wright TC Jr, Sharma A, Wright TL, Behrens CM. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol. 2011; 12:880–890. PMID: 21865084.

6. Satoh T, Matsumoto K, Fujii T, Sato O, Gemma N, Onuki M, et al. Rapid genotyping of carcinogenic human papillomavirus by loop-mediated isothermal amplification using a new automated DNA test (Clinichip HPVTM). J Virol Methods. 2013; 188:83–93. PMID: 23219807.
7. Herraez-Hernandez E, Alvarez-Perez M, Navarro-Bustos G, Esquivias J, Alonso S, Aneiros-Fernandez J, et al. HPV Direct Flow CHIP: a new human papillomavirus genotyping method based on direct PCR from crude-cell extracts. J Virol Methods. 2013; 193:9–17. PMID: 23680093.

8. Tao P, Zheng W, Wang Y, Bian ML. Sensitive HPV genotyping based on the flow-through hybridization and gene chip. J Biomed Biotechnol. 2012; 2012:938780. PMID: 23193367.

9. Hwang Y, Lee M. Comparison of the AdvanSure human papillomavirus screening real-time PCR, the Abbott RealTime High Risk human papillomavirus test, and the Hybrid Capture human papillomavirus DNA test for the detection of human papillomavirus. Ann Lab Med. 2012; 32:201–205. PMID: 22563555.

10. Song HJ, Lee JW, Kim BG, Song SY, Bae DS, Kim DS. Comparison of the performance of the PANArray™ HPV test and DNA chip test for genotyping of human papillomavirus in cervical swabs. BioChip J. 2010; 4:167–172.

11. An H, Song KS, Nimse SB, Kim J, Nguyen V, Ta VT, et al. HPV 9G DNA chip: 100% clinical sensitivity and specificity. J Clin Microbiol. 2012; 50:562–568. PMID: 22170909.

12. Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005; 32(Suppl 1):S16–S24. PMID: 15753008.

13. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009; 10:321–322. PMID: 19350698.
14. Arney A, Bennett KM. Molecular diagnositics of human papillomavirus. Lab Med. 2010; 41:523–530.
15. Lee GY, Kim SM, Rim SY, Choi HS, Park CS, Nam JH. Human papillomavirus (HPV) genotyping by HPV DNA chip in cervical cancer and precancerous lesions. Int J Gynecol Cancer. 2005; 15:81–87. PMID: 15670301.

16. Um TH, Lee EH, Chi HS, Kim JW, Hong YJ, Cha YJ. Comparison of HPV genotyping assays and Hybrid Capture 2 for detection of high-risk HPV in cervical specimens. Ann Clin Lab Sci. 2011; 41:48–55. PMID: 21325255.
Table 2
Concordance rates, Kappa, and PABAK values between HPVDNAChip and GenoBlot assays in each group
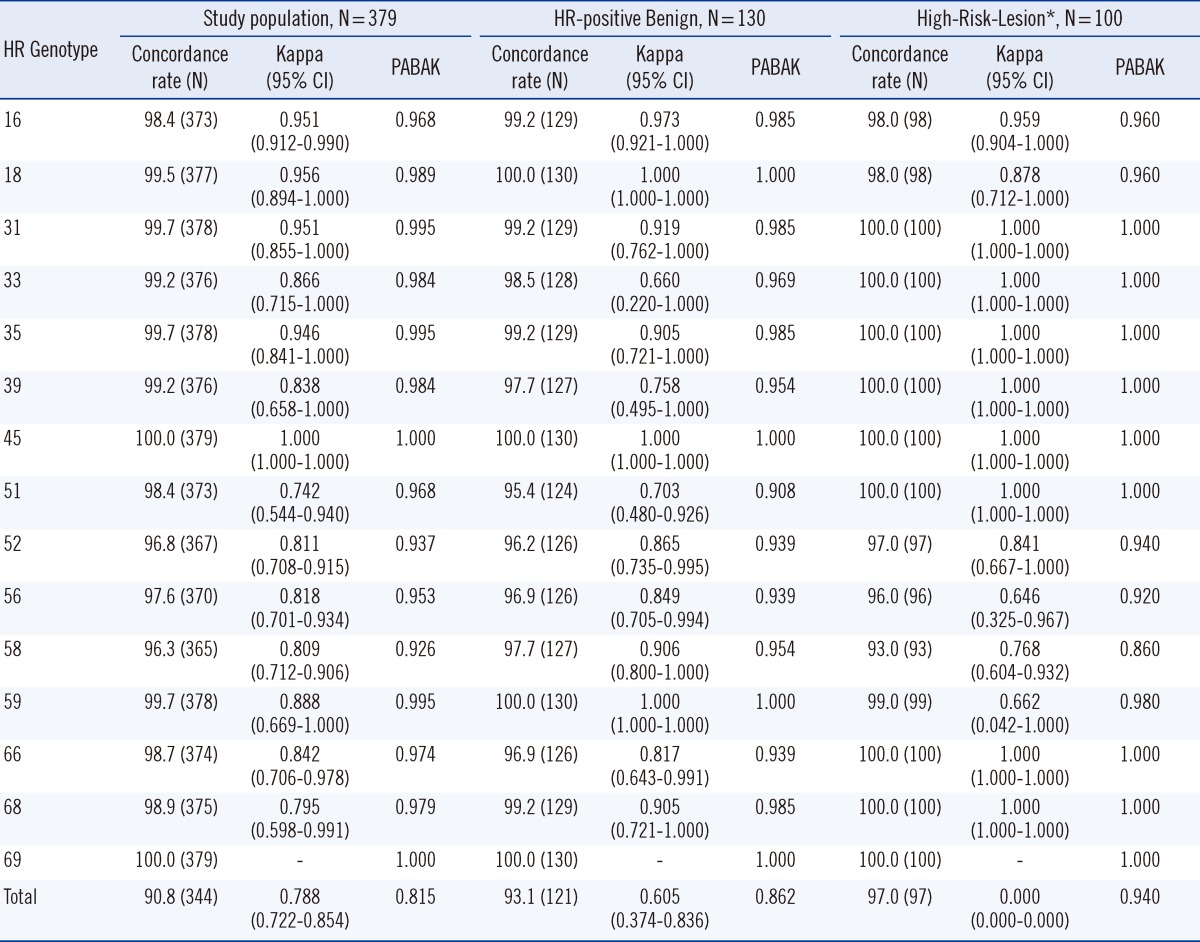
Table 4
Specificity and sensitivity of HPVDNAChip and GenoBlot assays for High-Risk-Lesion* diagnosis
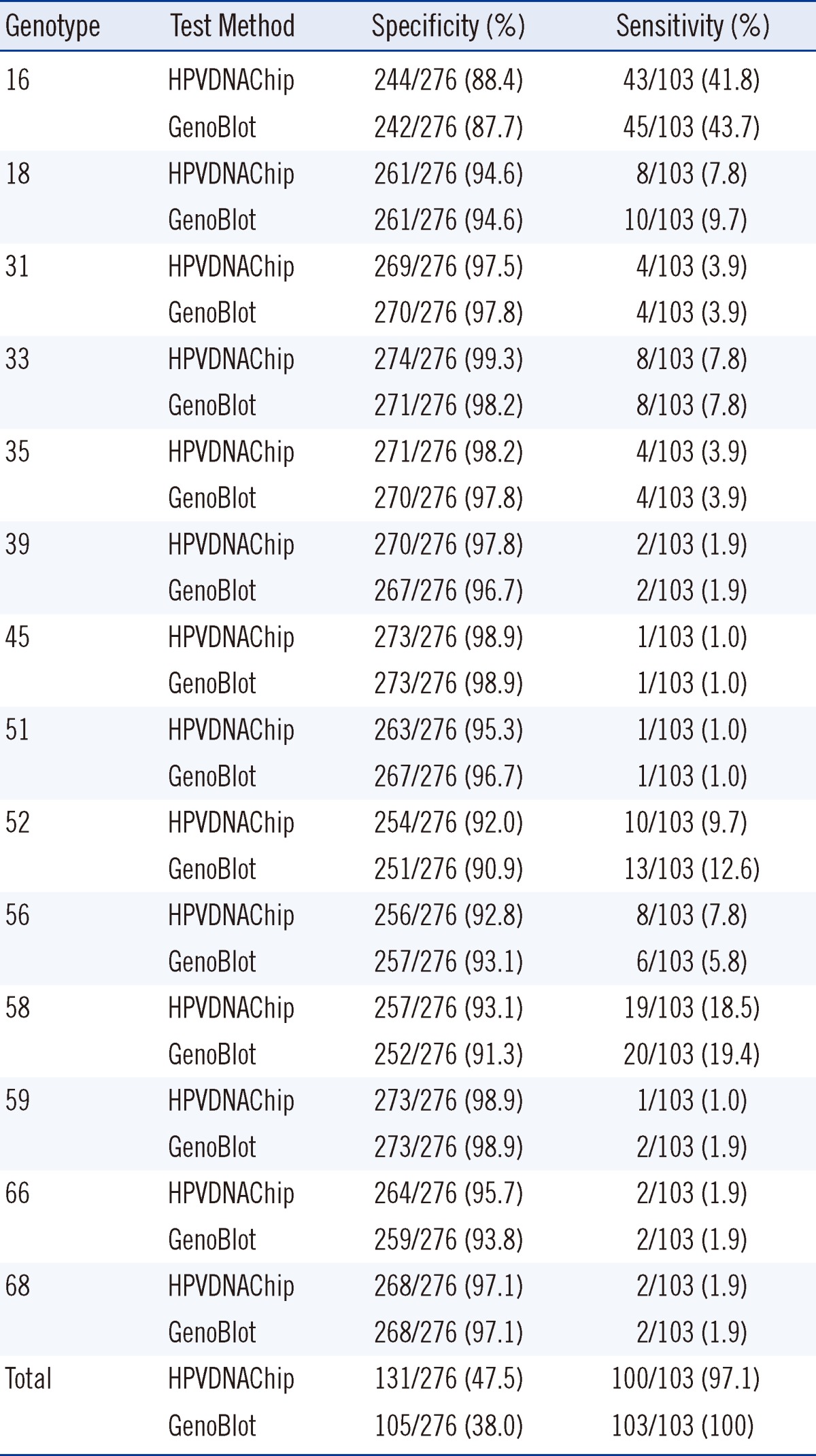
Table 5
Positive rate for each high-risk genotype by HPVDNAChip and GenoBlot assays in High-Risk-Lesion* Preoperative specimens during diagnosis vs. postoperative follow-up specimens




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download