Abstract
Background
Genetic abnormalities in adult AML are caused most frequently by somatic mutations in exon 12 of the NPM1 gene, which is observed in approximately 35% of AML patients and up to 60% of patients with cytogenetically normal AML (CN-AML).
Methods
We performed mutational analysis, including fragment analysis and direct sequencing of exon 12 of the NPM1 gene, on 83 AML patients to characterize the NPM1 mutations completely.
Results
In this study, NPM1 mutations were identified in 19 (22.9%) of the 83 AML patients and in 12 (42.9%) of the 28 CN-AML patients. Among the 19 patients with NPM1 mutations, type A NPM1 mutations were identified in 16 (84.2%) patients, whereas non-A type NPM1 mutations were observed in 3 (15.8%) patients. Two of the 3 non-A type NPM1 mutations were novel: c.867_868insAAAC and c.869_873indelCTTTAGCCC. These 2 novel mutant proteins display a nuclear export signal motif (L-xxx-L-xx-V-x-L) less frequently and exhibit a mutation at tryptophan 290 that disrupts the nucleolar localization signal.
Nucleophosmin (NPM) is a 294-amino acid phosphoprotein found at high levels in the granular regions of the nucleolus, and it is shuttled continuously between the nucleus and cytoplasm. This protein plays a key role in ribosome biogenesis, centrosome duplication, genomic stability, cell cycle progression, and apoptosis [1]. Depending on its dosage and level of expression, NPM is oncogenic or tumor-suppressing [2].
Recently, mutations in exon 12 of the NPM1 gene have been identified as the underlying genetic lesions in a large, distinct subgroup of adult patients with AML. In 2005, Falini et al. reported that NPM1 mutations on exon 12 resulted in aberrant cytoplasmic localization of NPM (NPMc+) in leukemic blasts in approximately 35% of adults with AML [3]; thus, NPM1 represents one of the most frequently mutated genes in AML [4]. Moreover, NPM1 gene mutations appear in approximately 60% of patients who present with cytogenetically normal AML (CN-AML) [3], and mutation status is related to treatment responsiveness and prognosis [5].
Wild-type NPM contains 2 nuclear export signal (NES) motifs, one within residues 94-102 and the other at the N terminus within amino acids 42-61 [6]. Wild-type NPM also contains a nucleolar localization signal (NLS) at its C terminus, which exports NPM from the cytoplasm to the nucleoplasm and finally to the nucleolus via its nucleolar-binding domains [7]. Although the role of NPM1 mutation in leukemogenesis has not been elucidated completely, the majority of NPM1 exon 12 mutations encode mutant proteins that have both a novel NES motif inserted at the C terminus and a disrupted NLS due to mutated tryptophan residues 288 and 290 (or 290 alone) [8-10]. The most common mutation in NPMc+ patients is type A, which duplicates a TCTG tetranucleotide in the reference sequence at 956-959 and accounts for 75-80% of adult AML with NPM1 mutations [11]. Recently, several studies have suggested that non-A type NPM1 mutations may function as prognostic factors for poor clinical outcomes [11, 12]. Therefore, it may be important to identify and characterize NPM1 mutations at the nucleotide and amino acid levels. In this study, we present the mutational spectrum of the NPM1 gene in Korean AML patients, which was directly sequenced.
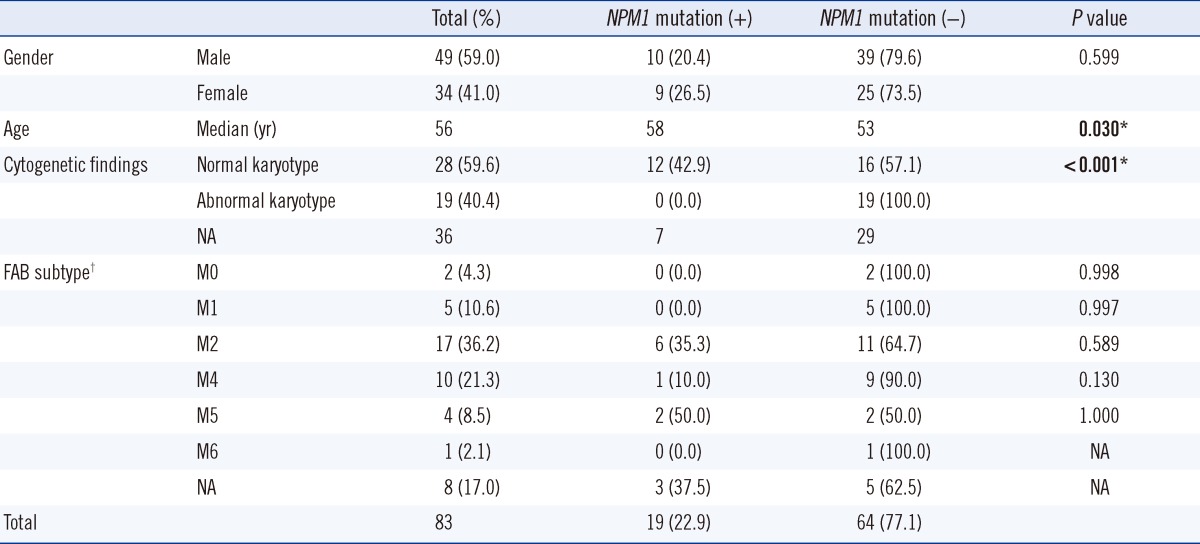
A total of 83 patients who were diagnosed with AML initially between October 2008 and September 2011 were included in the study. Their initial bone marrow or peripheral blood samples were collected for use in the study. Patients consisted of 49 men and 34 women; the median age was 56 yr (range, 16-86 yr). Cytogenetic information was available for 47 patients, and the French-American-British (FAB) subtype was available for 39 of those patients (Table 1). The study protocol was approved by the institutional review board of our hospital.
All 83 patient samples were subjected to fragment analysis and direct sequencing. Genomic DNA was extracted from bone marrow or peripheral blood using the PureGene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA). The NPM1 mutations were screened by fragment analysis of exon 12 of the NPM1 gene. The DNA region of interest was amplified using the following primers: 5'-6FAM-GGCCATATGGGTCTCTGTTC-3' and 5'-AACACGGTAGGGAAAGTTCTCA-3'. To determine the fragment size, amplified fragments and the appropriate size standards were detected using an ABI PRISM 3730xl genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Direct sequencing of exon 12 of the NPM1 gene was carried out by analysis of the amplified products by capillary electrophoresis using the ABI PRISM 3730xl genetic analyzer (Applied Biosystems). Sequencing reactions were prepared according to the manufacturer's instructions (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit; Applied Biosystems).
Mann-Whitney's U-test and Fisher's exact test were applied to determine the significance of the association between NPM1 mutations and other discrete variables among the patient subgroups. P values <0.05 were considered significant. All analyses were performed using SPSS Statistics 19 software (SPSS, Chicago, IL, USA).
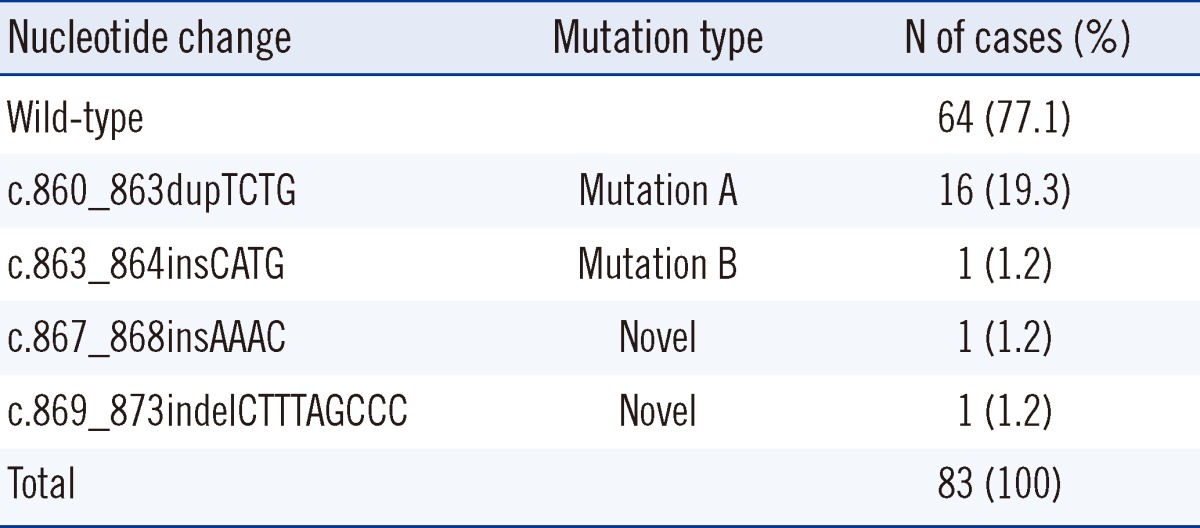
NPM1 gene mutations were identified in 19 of the 83 AML patients (22.9%) (Table 2). Of these 19 patients, type A mutation (c.860_863dupTCTG) was identified in 16 patients (84.2%), and type B mutation (c.863_864insCATG) was identified in 1 patient (5.3%). Interestingly, 2 novel NPM1 mutations were identified in the remaining 2 patients: c.867_868insAAAC (Patient 1) and c.869_873indelCTTTAGCCC (Patient 2) (Fig. 1).
Patient 1 was a 70-yr-old man with fever, myalgia, cough, and sputum. The results of his initial complete blood cell count (CBC) were as follows: hemoglobin, 8.0 g/dL; leukocytes, 10.1×109/L; platelets, 577×109/L. He exhibited hypercellular bone marrow that consisted of 63% blasts, which were positive for myeloperoxidase (MPO), CD13, CD33, and CD34 by flow cytometric analysis. The patient was diagnosed with AML M2, and the karyotype in all 20 metaphase cells analyzed was 46,XY. After treatment with re-induction chemotherapy, he achieved remission. Patient 2 was a 55-yr-old man who had experienced chest discomfort and dyspnea. The results of his initial CBC were as follows: hemoglobin, 3.9 g/dL; leukocytes, 53.0×109/L with 45% blasts; and platelets, 91×109/L. His bone marrow was replaced by 84.2% blasts. Flow cytometric analyses indicated that the blasts were positive for MPO, CD13, CD33, CD34, CD45, and CD117. The patient was diagnosed with AML M2, and the karyotype in all 11 metaphase cells was 46,XY. He also achieved remission after treatment with re-induction chemotherapy.
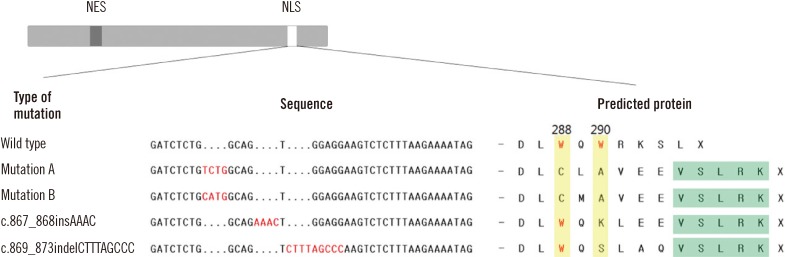
We predicted that these 2 mutations would lead to the synthesis of truncated forms of amino acids p.Trp290Lysfs*9 and p.Trp290Serfs*9, respectively (Fig. 2). The predicted amino acid sequences were DLWQKLEEVSLRK for Patient 1 and DLWQSLAQVSLRK for Patient 2.
Among the 47 AML patients whose karyotypes were available, 28 exhibited normal karyotypes (CN-AML), and the remaining 19 were abnormal. The number of NPM1 mutations detected in CN-AML (12/28, 42.9%) were higher than that in AML patients with abnormal karyotypes (0/19, 0.0%; P<0.001) (Table 1). With regard to FAB subtype of AML, NPM1 mutations were found more frequently in M2 (35.3%) and M5 (50.0%) than in the other subtypes of AML. Furthermore, older patients exhibited a higher incidence of NPM1 mutations (median age of patients with NPM1 versus without NPM1 mutation, 58 vs. 53 yr, P=0.030). NPM1 mutations were found more frequently in female patients (26.5%) than in male patients (20.4%), but this difference was not significant (P=0.599).
This study demonstrates similar NPM1 mutation rates in a CN-AML group (42.9%) compared to previous studies in the Korean population (35.3-53.7%) [11-13]. However, the rate determined here is lower than that of a European population (61.7%) [3]. NPM1 mutations are detected more often in AML M5 [14]. Therefore, in the present study, the differences in the clinical make-up of the study population, including a relatively lower incidence of AML M5, may have influenced the observed mutation rate in CN-AML.
Our study showed significantly higher numbers of NPM1 mutations in patients with normal karyotypes than that in patients with abnormal karyotypes (42.9 vs. 0.0%; P<0.001). This finding supports previous results that showed mutations in exon 12 of the NPM1 gene are the underlying genetic lesion in a distinct and large subgroup and that these mutations play a critical role in leukemogenesis in adult CN-AML [15]. It is noteworthy that none of the patients with karyotype abnormalities presented with NPM1 mutations. The mutation rate in the abnormal karyotype group was lower than that reported in previous studies [16-19]. In addition, our results suggest that an association exists between NPM1 mutation status and age at the time of diagnosis, which is consistent with a number of previous studies [14, 18, 19].
While most NPM1 mutations (including mutations A and B) have the NES motif L-xxx-V-xx-V-x-L, 2 novel NPM1 mutations that were identified in this study have a rare NES motif L-xxx-L-xx-V-x-L [4, 8]. The common NES motif requires the loss of both tryptophan 288 and 290 to be transported out of the nucleus efficiently. However, these novel mutations show a tryptophan loss at codon 290 only. NPM mutant proteins that retained tryptophan 288 accounted for 31% of all mutants in AML patients, while tryptophan 290 was mutated in all the NPM1 mutations identified [9]. We infer that tryptophan 290 may be more critical to nucleolar localization; thus, a single mutation at tryptophan 290 may be sufficiently relevant to AML pathogenesis. NPM can be exported efficiently despite the retention of tryptophan 288, but it is uncertain whether the novel NES motif and the disruption of the NLS can affect disease progression or prognosis [7, 9, 20].
Two recent studies suggested that a type A mutant impacted patient prognosis favorably [4, 21]. Another study showed that overall survival was shorter in non-A type mutants than that in patients with either no mutations or type A mutations of NPM1 [11]. Therefore, determination of the type of mutation in a patient may be relevant to predicting the prognosis. However, some available commercial kits or routine diagnostic methods designed to detect specific mutations (e.g., mutation A, B, or D) based on technologies like real-time PCR or fragment analysis may fail to identify other rare or novel NPM1 mutations.
In summary, we report 2 novel NPM1 mutations that were identified in AML patients. Although the prognostic implications of these novel mutations require further investigation, the present study suggests that novel NPM1 mutations may not be so rare. Supplementary sequence analysis and conventional targeted mutational analysis are required to detect non-A types of NPM1 mutations.
References
1. Eirín-López JM, Frehlick LJ, Ausió J. Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones. Genetics. 2006; 173:1835–1850. PMID: 16751661.
2. Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006; 6:493–505. PMID: 16794633.

3. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005; 352:254–266. PMID: 15659725.

4. Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. 2009; 27:171–181. PMID: 19569254.

5. Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005; 106:3740–3746. PMID: 16051734.
6. Yu Y, Maggi LB Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol Cell Biol. 2006; 26:3798–3809. PMID: 16648475.

7. Nishimura Y, Ohkubo T, Furuichi Y, Umekawa H. Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci Biotechnol Biochem. 2002; 66:2239–2242. PMID: 12450141.

8. Falini B, Nicoletti I, Martelli MF, Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood. 2007; 109:874–885. PMID: 17008539.

9. Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006; 107:4514–4523. PMID: 16455950.

10. Mariano AR, Colombo E, Luzi L, Martinelli P, Volorio S, Bernard L, et al. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene. 2006; 25:4376–4380. PMID: 16501600.

11. Koh Y, Park J, Bae EK, Ahn KS, Kim I, Bang SM, et al. Non-A type nucleophosmin 1 gene mutation predicts poor clinical outcome in de novo adult acute myeloid leukemia: differential clinical importance of NPM1 mutation according to subtype. Int J Hematol. 2009; 90:1–5. PMID: 19484332.

12. Park BG, Chi HS, Park SJ, Min SK, Jang S, Park CJ, et al. Clinical implications of non-A-type NPM1 and FLT3 mutations in patients with normal karyotype acute myeloid leukemia. Acta Haematol. 2012; 127:63–71. PMID: 22104247.

13. Kim YK, Kim HN, Lee SR, Ahn JS, Yang DH, Lee JJ, et al. Prognostic significance of nucleophosmin mutations and FLT3 internal tandem duplication in adult patients with cytogenetically normal acute myeloid leukemia. Korean J Hematol. 2010; 45:36–45. PMID: 21120161.

14. Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011; 117:1109–1120. PMID: 21030560.

15. Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009; 23:1731–1743. PMID: 19516275.

16. Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005; 106:3747–3754. PMID: 16109776.

17. Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006; 107:4011–4020. PMID: 16455956.

18. Ahmad F, Mandava S, Das BR. Mutations of NPM1 gene in de novo acute myeloid leukaemia: determination of incidence, distribution pattern and identification of two novel mutations in Indian population. Hematol Oncol. 2009; 27:90–97. PMID: 19365794.
19. Yan L, Chen S, Liang J, Feng Y, Cen J, He J, et al. Analysis of NPM1 gene mutations in Chinese adults with acute myeloid leukemia. Int J Hematol. 2007; 86:143–146. PMID: 17875528.

20. Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005; 7:823–830. PMID: 16041368.

21. Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009; 21:573–581. PMID: 19770764.

Fig. 1
Sequence analysis of the 2 novel NPM1 mutations. (A) c.867_868insAAAC, (B) c.869_873indelCTTTAGCCC, and (C) the wild-type NPM1 sequence.
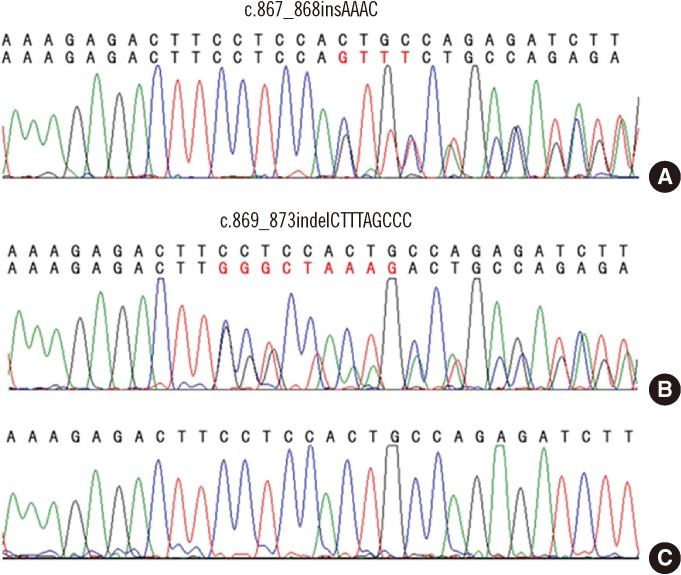
Fig. 2
Nucleotide and amino acid sequences of the 2 novel NPM1 mutations. Wild type, type A (c.860_863dupTCTG), and type B mutations (c.863_864insCATG) are presented here. Tryptophans 288 and 290 (yellow background) are replaced by another amino acid in mutations A and B, whereas tryptophan 288 is retained in the 2 novel mutations identified in this study. The latter part of hydrophobic residue-rich NES motif (green background) is common to all types of mutation.
Abbreviations: NES, nuclear export signal; NLS, nucleolar localization signal.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download