Weissella confusa is a catalase-negative and gram-positive coccobacillus that is intrinsically resistant to vancomycin [1]. This species can cause infections, such as bacteremia, endocarditis, and abscess in immunocompromised patients [2-7]. Until now, reports on infections caused by W. confusa have been limited as this species has been frequently misidentified as Lactobacillus or Leuconostoc when identified using commercial kits [8]. Herein, we report the second case of W. confusa bacteremia in an immunocompetent patient from Korea, who had intramural hematomas of the aorta.
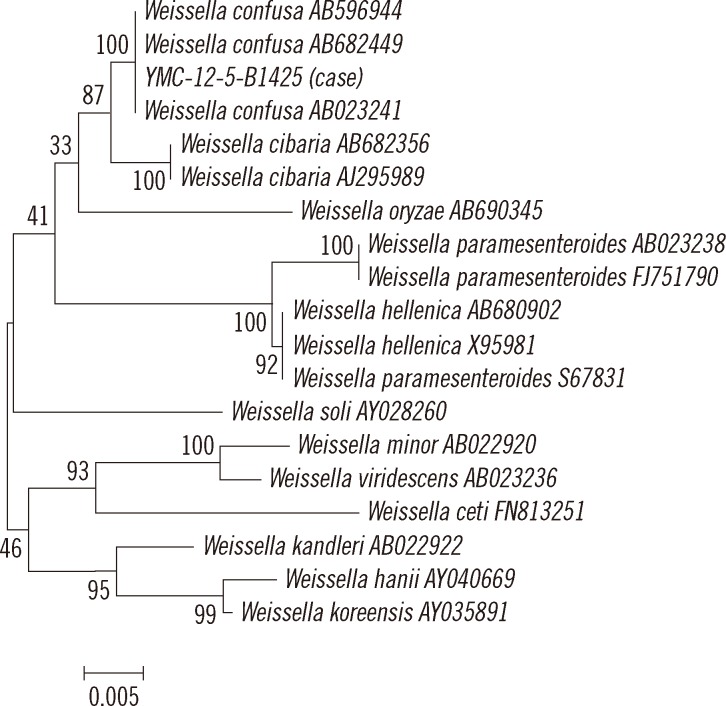
A 60-yr-old woman with no history of illness, except hypertension, was admitted to the hospital complaining of abdominal discomfort, distension, and symptoms of gastroesophageal reflux disease during the previous several days. Initial vital signs were: blood pressure of 110/70 mm Hg, pulse rate of 86/min, respiratory rate of 20/min, and body temperature of 37.1℃. Laboratory investigations showed Hb 11.2 g/dL, leukocyte count 8.61×109/L, platelet count 188×109/L, AST/ALT 65/123 IU/L, alkaline phosphatase 151 IU/L, and elevated C-reactive protein level 155.7 mg/dL. Chest computerized tomography revealed intramural hematomas over the entire thoracic and proximal abdominal aorta. On the day after admission, the patient presented with fever (38.3℃). Blood samples for culture were inoculated into 3 sets of Bact/ALERT FA and FN bottles (bioMérieux Inc., Durham, NC, USA). Gram-positive coccobacilli from 1 pair of FA and FN bottles were isolated. After incubation at 37℃ with 5% CO2 for 24 hr, small-sized α-hemolytic colonies were observed on sheep blood agar (Fig. 1). The organism was identified as gram-positive coccobacilli (Fig. 2). It was negative for oxidase, catalase, pyrrolidonyl-arylamidase, and leucine aminopeptidase and had a positive bile-esculin reaction. The minimum inhibitory concentration of vancomycin was determined to be >256 µg/mL by using E-test strip (AB bioMérieux, Solna, Sweden). The isolate was initially assumed to be a Leuconostoc species because of its biochemical characterizations and vancomycin resistance. It was subsequently identified as Streptococcus equinus and Aerococcus viridans according to the gram-positive identification card of the Vitek 1 and the gram-positive card of the Vitek 2 systems (bioMérieux Inc.), respectively. For further identification, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, VITEK MS system, bioMérieux Inc.) was used. Mass spectra were acquired by using the Axima Assurance mass spectrometer (Shimadzu Corporation, Kyoto, Japan) and analyzed with the VITEK MS IVD v1 database (bioMérieux Inc.). The strain was identified as W. confusa, which has a similar biochemical profile to that of Leuconostoc [1], with a confidence value of 99.9%. In addition, we performed 16S rRNA gene sequencing of a PCR product sized about 1.5 kb by using the universal primers 8F (5'-AGA GTT TGA TCC TGG CTC AG-3') and 1541R (5'-AAG GAG GTG ATC CAG CCG CA-3') and compared the obtained sequence with the EzTaxon database [9] and the bioinformatic bacterial identification database [10]. When the sequence was submitted to the ExTaxon database, the isolate revealed 99.66% similarity (1,471/1,476 bp) with W. confusa. The next closest matches were W. cibaria, W. oryzae, W. viridescens, and W. paramesenteroides with similarities of 99.06% (1,480/1,494 bp), 96.66% (1,417/1,466 bp), 96.28% (1,422/1,477 bp), and 96.18% (1,434/1,491 bp), respectively. In the bioinformatic bacterial identification database search, the isolate exhibited 99.66% similarity (1,467/1,472 bp) with W. confusa AB023241 followed by W. confusa AB682449, W. confusa AB596944, W. cibaria AB682356, and W. cibaria AJ295989 with similarities of 99.60% (1,485/1,491 bp), 99.60% (1,485/1,491 bp), 98.99% (1,476/1,491 bp), and 98.86% (1,474/1,491 bp), respectively. A phylogenetic tree, which was constructed with the neighbor-joining method by using Molecular Evolutionary Genetics Analysis (MEGA) software version 5.05 [11], revealed this isolate to be W. confusa (Fig. 3).
Empirical therapy with ceftriaxone was initiated after blood culture submission. In spite of the treatment, fever persisted for 3 days and the C-reactive protein level did not decrease. The ceftriaxone treatment was changed to teicoplanin plus piperacillin-tazobactam after isolation of gram-positive coccobacilli from the blood culture. Fever and other symptoms subsided with the new regimen. All subsequent blood cultures were negative, and the patient was discharged on the 9th hospital day with an uneventful recovery.
On the basis of the 16S rRNA gene sequence analysis, Leuconostoc paramesenteroides and related species among the catalase-negative, vancomycin-resistant, gram-positive cocci were reclassified into a new genus, Weissella, in 1993 [12]. There are 16 species in the Weissella genus, but only W. confusa (previously Lactobacillus confusus) and W. cibaria have been isolated from clinical specimens. While W. confusa infections are not common, a case of infectious endocarditis caused by W. confusa in Korea has been reported [5]. The characteristics of 10 bacteremia cases of W. confusa in Taiwan were reviewed [8]. Among the 43 isolates, which were identified as either Lactobacillus or Leuconostoc species using commercial identification systems, 10 isolates were confirmed as W. confusa using 16S rRNA gene sequencing, and all affected patients had complex medical conditions and/or an immunocompromised status. In additional reports, W. confusa isolates could not be identified using conventional identification systems except by 16S rRNA gene sequence analysis, and most patients were immunocompromised [2-4, 6].
When clinical isolates cannot be identified by using conventional methods, 16S rRNA gene sequence analysis can be useful [13]. However, routine application of 16S rRNA gene sequencing has some limitations since it is relatively expensive and time-consuming. On the other hand, MALDI-TOF MS can identify bacteria within a few minutes using colonies grown on culture plates, and it can reduce the turnaround time and the cost of expendables [14]. During the past several years, there have been numerous reports demonstrating the accuracy and reliability of the MALDI-TOF MS system [15-18].
In this case, gram-positive coccobacilli from the blood culture was initially suspected to be a Leuconostoc species due to its biochemical characteristics, i.e., vancomycin resistance and negative results with catalase, pyrrolidonyl-arylamidase, and leucine aminopeptidase [1]. However, the isolate was identified as W. confusa using MALDI-TOF MS, which was confirmed by 16S rRNA gene sequence analysis.
W. confusa is taxonomically very closely related to W. cibaria, but can be differentiated form W. cibaria because of differences in the biochemical characteristics, e.g., it is positive for fermentation of galatose and xylose and negative for fermentation of arabinose [19]. However, both species cannot be differentiated by using the VITEK MS system alone because W. cibaria is not included in the VITEK MS IVD v1 database. Therefore, 16S rRNA gene sequence analysis and/or additional biochemical tests are needed for the accurate identification.
In blood cultures, contaminations of normal skin flora occur often and include coagulase-negative staphylococci, Corynebacterium species, Propionibacterium acnes, Micrococcus species, and viridans group streptococci [20]. Unless the isolate grows in multiple blood culture sets, identification to the species level may not be warranted. Although the W. confusa isolate was cultured from only 1 set in this study and the patient was not immunocompromised, we regarded the strain as the true pathogen because the C-reactive protein level was markedly elevated. Furthermore, fever persisted despite treatment with ceftriaxone and subsided after changing the regimen to teicoplanin plus piperacillin-tazobactam, which is one of the regimens for the treatment of W. confusa infections.
The standard methods and interpretation criteria of antimicrobial susceptibilities for W. confusa are not yet established. In a recent study [8], amoxicillin-clavulanic acid, ampicillin-sulbactam, piperacillin-tazobactam, daptomycin, moxifloxacin, doripenem, and tigecycline revealed relatively good in vitro activity and were recommended for the treatment of W. confusa infections.
In summary, we described a second case of bacteremia caused by W. confusa in an immunocompetent host. The isolate was not identified when commercial identification kits were used but could be identified using 16S rRNA gene sequence analysis and a MALDI-TOF MS system. When blood cultures show growth of catalase-negative, gram-positive coccobacilli with negative pyrrolidonyl-arylamidase and leucine aminopeptidase reaction, clinical microbiologists should be aware of the possibility of W. confusa infection.
References
1. Ruoff KL. Aerococcus, Abiotrophia, and other aerobic catalase-negative, Gram-positive cocci. In : Versalovic J, editor. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press;2011. p. 365–376.
2. Salimnia H, Alangaden GJ, Bharadwaj R, Painter TM, Chandrasekar PH, Fairfax MR. Weissella confusa: an unexpected cause of vancomycin-resistant gram-positive bacteremia in immunocompromised hosts. Transpl Infect Dis. 2011; 13:294–298. PMID: 21156010.
3. Kumar A, Augustine D, Sudhindran S, Kurian AM, Dinesh KR, Karim S, et al. Weissella confusa: a rare cause of vancomycin-resistant Gram-positive bacteraemia. J Med Microbiol. 2011; 60:1539–1541. PMID: 21596906.
4. Harlan NP, Kempker RR, Parekh SM, Burd EM, Kuhar DT. Weissella confusa bacteremia in a liver transplant patient with hepatic artery thrombosis. Transpl Infect Dis. 2011; 13:290–293. PMID: 21504525.
5. Shin JH, Kim DI, Kim HR, Kim DS, Kook JK, Lee JN. Severe infective endocarditis of native valves caused by Weissella confusa detected incidentally on echocardiography. J Infect. 2007; 54:e149–e151. PMID: 17052757.
6. Olano A, Chua J, Schroeder S, Minari A, La Salvia M, Hall G. Weissella confusa (basonym: Lactobacillus confusus) bacteremia: a case report. J Clin Microbiol. 2001; 39:1604–1607. PMID: 11283096.
7. Bantar CE, Relloso S, Castell FR, Smayevsky J, Bianchini HM. Abscess caused by vancomycin-resistant Lactobacillus confusus. J Clin Microbiol. 1991; 29:2063–2064. PMID: 1774335.
8. Lee MR, Huang YT, Liao CH, Lai CC, Lee PI, Hsueh PR. Bacteraemia caused by Weissella confusa at a university hospital in Taiwan, 1997-2007. Clin Microbiol Infect. 2011; 17:1226–1231. PMID: 21040157.
9. Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007; 57:2259–2261. PMID: 17911292.

10. Devulder G, Perrière G, Baty F, Flandrois JP. BIBI, a bioinformatics bacterial identification tool. J Clin Microbiol. 2003; 41:1785–1787. PMID: 12682188.

11. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739. PMID: 21546353.

12. Collins MD, Samelis J, Metaxopoulos J, Wallbanks S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J Appl Bacteriol. 1993; 75:595–603. PMID: 8294308.
13. Park KJ, Park KS, Choi SH, Kim YJ, Ki CS, Kang IS, et al. Haemophilus parainfluenzae infective endocarditis confirmed by 16S rRNA sequence analysis from culture negative tissue. Korean J Clin Microbiol. 2012; 15:139–142.
14. Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol. 2012; 93:965–974. PMID: 22198716.

15. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010; 48:1549–1554. PMID: 20220166.

16. Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, et al. Rapid identification and typing of listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol. 2008; 74:5402–5407. PMID: 18606788.
17. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008; 46:1946–1954. PMID: 18400920.

18. Kim M, Kwon MJ, Chung HS, Lee Y, Yong D, Jeong SH, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of aerobic bacteria in a clinical microbiology laboratory. Korean J Clin Microbiol. 2012; 15:60–66.

19. Björkroth KJ, Schillinger U, Geisen R, Weiss N, Hoste B, Holzapfel WH, et al. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int J Syst Evol Microbiol. 2002; 52:141–148. PMID: 11837296.
20. Hall KK. Updated review of blood culture contamination. Clin Microbiol Rev. 2006; 19:788–802. PMID: 17041144.

Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download