Abstract
Background
This study aimed to evaluate the prevalence of Mycoplasma pneumoniae in primary and tertiary care hospitals and its macrolide resistance rate.
Methods
Nasopharyngeal swabs were collected from 195 pediatric patients in primary and tertiary care hospitals from October to November 2010. The AccuPower MP real-time PCR kit (Bioneer, Korea) was used for the detection of M. pneumoniae. Direct amplicon sequencing was performed to detect point mutations conferring resistance to macrolides in the 23S rRNA gene.
Results
Among the 195 specimens, 17 (8.7%) were M. pneumoniae positive, and 3 of the strains (17.6%) obtained from these 17 specimens displayed the A2063G mutation in 23S rRNA. Three macrolide-resistant M. pneumoniae isolates were isolated from patients hospitalized at the primary care hospital. The positive rates of M. pneumoniae for the primary and tertiary care hospitals were 12.1% (15/124) and 2.8% (2/71), respectively (P=0.033).
Conclusions
The positive rate of M. pneumoniae in the primary care hospital was higher than that in the tertiary care hospital. Simultaneous detection of M. pneumoniae and macrolide-resistant mutation genes in the 23S rRNA by real-time PCR is needed for rapid diagnosis and therapy of M. pneumoniae infections.
Go to : 
Mycoplasma pneumoniae infections can involve both the upper and lower respiratory tracts in children and adults and occur both endemically and epidemically worldwide [1]. Although many M. pneumoniae infections result in mild, asymptomatic, and often self-limiting cases, up to 25% of those infected with M. pneumoniae may experience extra-pulmonary complications and/or require hospitalization due to severe pneumonia, especially children [1, 2]. M. pneumoniae can be transmitted through aerosols in typical outbreak settings, in which close physical contact occurs (e.g., home, school, military barracks, and dormitories). Therefore, proper choice of antibiotics, based on rapid and sensitive diagnostic methods, is essential for treating children with M. pneumoniae infections [3].
Among the several diagnostic methods for detecting M. pneumoniae in respiratory specimens, real-time PCR has emerged as a significant improvement for the rapid diagnosis of this pathogen. It has been shown that real-time PCR may be more useful during the early stages of the infection, probably because of the presence of a higher pathogen burden and because the specimen collection occurs prior to the antibiotic treatment [2]. Therefore, specimen collection and testing should be performed as early as possible.
Despite the advances in molecular technology, acute care facilities and primary care hospitals rarely perform molecular tests, because it requires specialized equipment and highly trained personnel, as well as longer processing time to obtain results. As a result, the actual incidence of M. pneumoniae infections among individuals seeking medical care for acute respiratory symptoms is more likely to be underestimated and may lead to inappropriate therapy, including unwarranted use of antibiotics or inappropriate antibiotic choice [4]. To the best of our knowledge, there has been no report describing the difference of detection rate and resistance rate of M. pneumoniae between primary and tertiary care hospitals using real-time PCR.
Macrolides are frequently used as empirical treatments for upper respiratory infections, because they are effective against major pathogens, such as β-hemolytic streptococci, Streptococcus pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and M. pneumoniae [5]. Particularly, school-age children with symptoms and signs of pneumonia are usually treated empirically with macrolide antibiotics, because M. pneumoniae is a leading cause of community-acquired pneumonia in this age group. Moreover, children are usually not treated with quinolones because of the risk of side effects [6]. However, macrolide-resistant M. pneumoniae has been detected in Japan, Italy, France, USA, and Denmark [3], and the detection rate reached 90% in China [7].
Herein, we analyzed the macrolide resistance rate and prevalence of M. pneumoniae in primary and tertiary care hospitals.
Go to : 
Between October and November 2010, 195 nasopharyngeal swabs were collected from pediatric patients treated at Wonju Severance Christian Hospital (WSCH) and Wonju Medical Center (WMC) in Wonju, Korea. WSCH, a tertiary care university hospital, is located about 1.7 km away from the WMC, which is a primary care hospital. Nasopharyngeal swabs were collected using flocked swabs and transported in universal transport medium (Copan Diagnostics, Corona, Italy). Nasopharyngeal swab specimens obtained at WMC were transported to WSCH within 24 hr of collection. Information regarding antibiotic use and the duration of antibiotic therapy before specimen collection was obtained.
M. pneumoniae DNA was extracted with the Exiprep genomic DNA kit (Bioneer, Daejeon, Korea). The AccuPower MP real-time PCR kit (Bioneer) was used for detecting M. pneumoniae according to the manufacturer's instructions. Briefly, M. pneumoniae-specific DNA regions were detected via the fluorescence of specific hydrolysis probes (5'-fluorescein carboxylic acid [FAM] and 3'-black hole quencher-1 [BHQ1]) during thermal cycling (Exicycler 96 Real-Time Quantitative Thermal Block, Bioneer). PCR was carried out using a PCR premix kit for the detection of M. pneumoniae (Bioneer). Five microliters of extracted DNA, positive control DNA, or distilled water (negative control) were added into individual premix tubes. The reaction mixture was then subjected to denaturation for 10 min at 95℃, then to 40 PCR cycles, each of 20 sec at 95℃ and 30 sec at 55℃, followed by a final cooling step of 1 min at 25℃. The CT (cycle threshold) value for which PCR was defined positive was less than 40.
A PCR assay followed by direct amplicon sequencing was developed to detect point mutations conferring resistance to macrolides in the V domain of the M. pneumoniae 23S rRNA gene. After we obtained the DNA sequence of M. pneumoniae from GenBank (NIH, Bethesda, MD, USA), we designed primers for detecting known M. pneumoniae single point mutations (A2063G, A2064G, T2611C, and C2617A). The forward primer (target gene: 23S rRNA 1,851-2,675 bp) was 5'-GAA GGT TAA AGA AGG AGG TTA GCG CAA-3', and the reverse primer was 5'-TCG GTC CTC TCG TAC TAG AAG CAA CA-3'. Amplification of the 23S rRNA gene of M. pneumoniae was performed using MyGenie 32 Thermal Block thermal cycler (Bioneer). PCR was carried out using 20 µL of the high fidelity PCR premix (Bioneer) with the following profile: denaturation for 5 min at 94℃; 35 cycles of 30 sec at 94℃, 30 sec at 55℃, and 1 min at 72℃; final extension at 10 min at 72℃; and cooling at 4℃. The amplified products were separated by electrophoresis on a 1.5% agarose gel and visualized under ultraviolet light. The amplicons were purified using gel purification (Bioneer) and directly sequenced on ABI Prism 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA).
Data were analyzed using IBM SPSS Statistics version 20 (SPSS Inc., Chicago, IL, USA). Statistical significance of differences in categorical variables, such as hospital size, age group, initial antibiotic therapy before PCR request, and sex, was determined by the Chi-square test or Fisher's exact test. Statistical significance was determined at the 0.05 level of a two-tailed test.
Go to : 
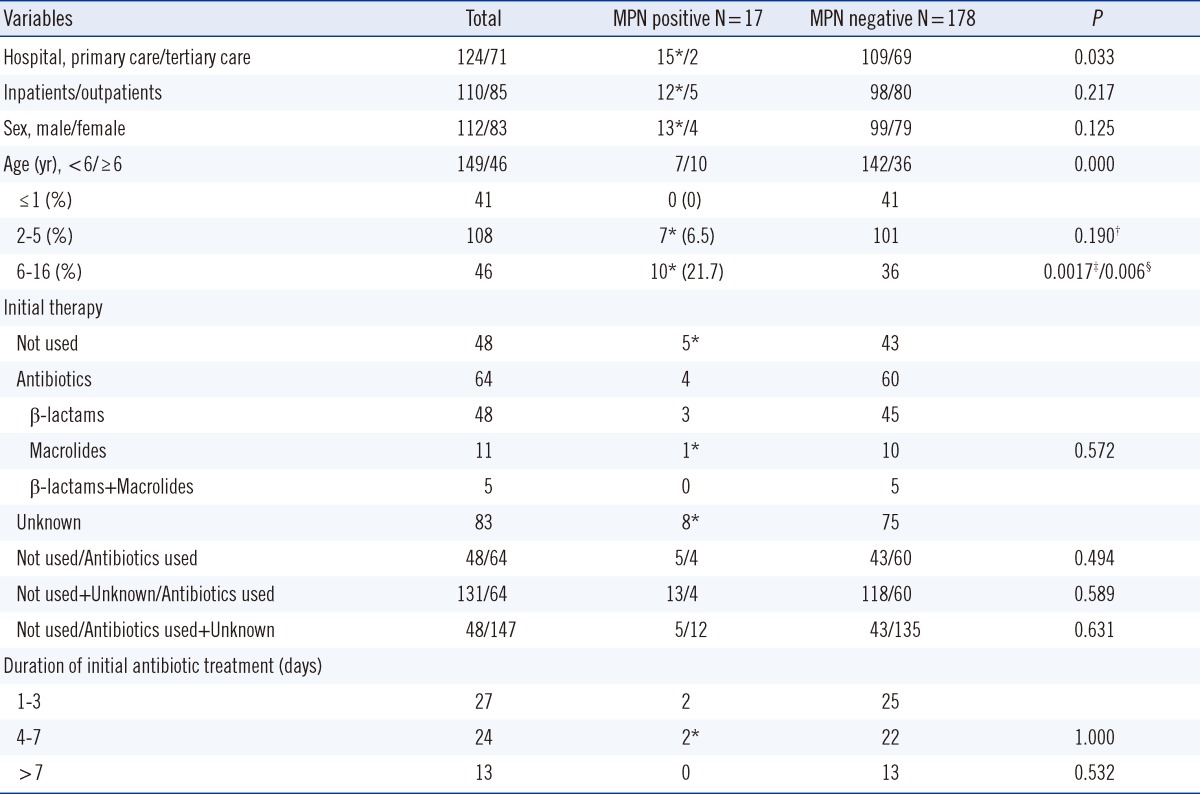
Among a total of 195 specimens, 17 (8.7%) were positive for M. pneumoniae. The positive rates of M. pneumoniae for the primary care hospital and tertiary care hospital were 12.1% (15/124) and 2.8% (2/71), respectively (P=0.033). The positive rates of M. pneumoniae according to age group were 0% (0/41) for ≤1 yr, 6.5% (7/108) for 2-5 yr, and 21.7% (10/46) for 6-16 yr. The use of antibiotics before specimen collection, the duration of initial antibiotic therapy, the ratio of inpatients to outpatients, and the ratio of male to female showed no statistical differences between the M. pneumoniae-positive and M. pneumoniae-negative groups. Among the 17 M. pneumoniae-positive cases, 3 strains (17.6%) displayed an A-to-G point mutation at nucleotide 2,063 in 23S rRNA. Three macrolide-resistant M. pneumoniae isolates were isolated from patients hospitalized at the primary care hospital (Table 1).
Go to : 
PCR seems to be a very sensitive and practical diagnostic tool for detecting acute M. pneumoniae infections [8]. However, the sensitivity of nucleic acid-based tests is usually affected by the type and quality of the respiratory specimen [9]. Although M. pneumoniae rarely colonizes the respiratory tract, it can persist for variable periods in the respiratory tract following infection that has resolved clinically with appropriate antimicrobial therapy [1]. For this reason, a combination of nucleic acid-based and serologic tests has been the most reliable approach for the diagnosis of M. pneumoniae infection.
In general, PCR-based technology has disadvantages, such as the possible contamination of laboratory PCR product due to repetitive testing and the requirement of post-PCR steps. Recently, more sensitive and specific real-time PCR methods targeting many genes of M. pneumoniae, such as the ATPase operon gene, p1 adhesin gene, RepMp1 gene, and CARDS toxin gene, have been developed in Europe and United States [10]. In Korea, PCR-based approaches, such as nested PCR or multiplex PCR, have mainly been used for M. pneumoniae detection [6, 9, 11]. Bioneer, a Korean company, has recently launched a real-time PCR kit for the detection of M. pneumoniae.
The appropriate specimen type for the molecular diagnosis of M. pneumoniae pneumonia remains controversial. M. pneumoniae was more abundant in sputum than in upper respiratory tract samples in clinical specimens. In the study evaluating specimen types for simultaneous detection of M. pneumoniae, C. pneumoniae, and L. pneumophila using multiplex PCR, the detection rate of M. pneumoniae from sputum samples was higher than those from nasopharyngeal swabs (9.2% vs. 3.7%) [9]. However, since only adult community-acquired pneumonia patients were included in that study, the difference in sensitivity between upper and lower respiratory tract specimens may be more pronounced than in the studies that included patients with both upper and lower respiratory tract infections of M. pneumoniae. Sasaki et al. [12] reported that positive PCR result for M. pneumoniae did not significantly differ between throat and nasopharyngeal specimens, but it correlated with the disease severity. Nasopharyngeal swab and sputum were considered as the preferred specimen types for younger children and adults, respectively.
The positive rate of M. pneumoniae varies depending on study period, age group, detection method, and specimen type. M. pneumoniae is endemic in the larger communities worldwide, but epidemics, lasting several months to years, periodically occur every 3 to 7 yr. Jung et al. [13] reported that the positive rate of M. pneumoniae in adult patients with pneumonia between October 2008 and April 2009 was 2.0% (2/100). Hong et al. [14] reported that the positive rate of M. pneumoniae in pediatric patients who were diagnosed as having lower respiratory tract infections from August 2010 to December 2011 was 7.2%. In this study, the overall positive rate for M. pneumoniae was 8.7%, with the positive rate in a primary care hospital higher than that of a tertiary care hospital (12.1% vs. 2.8%). This result suggests that delaying specimen collection after the symptoms onset is the major factor determining positive rate of M. pneumoniae.
The prevalence of macrolide-resistant M. pneumoniae (MRMP) has been reported to be relatively low in the United States and Europe: 0.9-2.9% in Denmark, 3% in Germany, 10% in France, 26% in Italy, and 32% in Israel [3]. Like neighboring Korea, China is now showing high MRMP prevalence, accounting for over 80% of M. pneumoniae in children and adults [7, 15]. In Japan, the prevalence of MRMP in children has increased rapidly from 5.0% in 2003 to 39% in 2008 and to 87.1% in 2011 [3]. MRMP has also been isolated from young adult patients with community-acquired pneumonia [16]. In Korea, on the basis of the study performed using 62 frozen-stocked nasopharyngeal aspirates specimens between 2000 and 2003 [17], 17 had M144V mutations in the ribosomal protein L4 and 1 specimen had an A2064G mutation in the V domain of the 23S rRNA gene of M. pneumoniae. In contrast, Yoo et al. [6] reported that the frequency of the A2063G mutation in the 23S rRNA gene of M. pneumoniae in 2011 was significantly higher in children than in adults (61.3% vs. 13.3%) and that there were no other known mutations such as A2063C, A2064G, or A2067G. In our study performed in 2010, the prevalence of MRMP was 17.6%. The prevention of more extensive outbreaks of MRMP infection requires more rapid diagnosis, because MRMP infection is significantly related to school age and patients with MRMP infections show a long duration of fever and require a long duration of antibiotic treatment [6].
Go to : 
Acknowledgements
We would like to thank Bioneer (Daejeon, Korea) for providing us with M. pneumoniae DNA extraction kits and AccuPower MP real-time PCR kit for this study.
Go to : 
References
1. Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004; 17:697–728. PMID: 15489344.
2. Winchell JM, Mitchell SL. Detection of Mycoplasma pneumoniae by real-time PCR. Methods Mol Biol. 2013; 943:149–158. PMID: 23104288.
3. Okada T, Morozumi M, Tajima T, Hasegawa M, Sakata H, Ohnari S, et al. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin Infect Dis. 2012; 55:1642–1649. PMID: 22972867.
4. Diaz MH, Winchell JM. Detection of Mycoplasma pneumoniae and Chlamydophila pneumoniae directly from respiratory clinical specimens using a rapid real-time polymerase chain reaction assay. Diagn Microbiol Infect Dis. 2012; 73:278–280. PMID: 22541789.
5. Yao JDC, Moellering RC. Antibacterial agents. In : Versalovic J, editor. Manual of clinical microbiology. 10th ed. Washington DC: ASM press;2011. p. 1043–1081.
6. Yoo SJ, Kim HB, Choi SH, Lee SO, Kim SH, Hong SB, et al. Differences in the frequency of 23S rRNA gene mutations in Mycoplasma pneumoniae between children and adults with community-acquired pneumonia: clinical impact of mutations conferring macrolide resistance. Antimicrob Agents Chemother. 2012; 56:6393–6396. PMID: 23006753.
7. Liu Y, Ye X, Zhang H, Xu X, Li W, Zhu D, et al. Characterization of macrolide resistance in Mycoplasma pneumoniae isolated from children in Shanghai, China. Diagn Microbiol Infect Dis. 2010; 67:355–358. PMID: 20638604.
8. Ferwerda A, Moll HA, de Groot R. Respiratory tract infections by Mycoplasma pneumoniae in children: a review of diagnostic and therapeutic measures. Eur J Pediatr. 2001; 160:483–491. PMID: 11548186.
9. Cho MC, Kim H, An D, Lee M, Noh SA, Kim MN, et al. Comparison of sputum and nasopharyngeal swab specimens for molecular diagnosis of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella pneumophila. Ann Lab Med. 2012; 32:133–138. PMID: 22389880.
10. Zhao F, Cao B, He LH, Yin YD, Tao XX, Song SF, et al. Evaluation of a new real-time PCR assay for detection of Mycoplasma pneumoniae in clinical specimens. Biomed Environ Sci. 2012; 25:77–81. PMID: 22424630.
11. Lim HK, Chung YH. Significance of polymerase chain reaction test for diagnosis of Mycoplasma pneumoniae pneumonia. J Korean Pediatr Soc. 1999; 42:173–179.
12. Sasaki A, Ouchi K, Makata H, Hashimoto K, Matsubara T, Furukawa S. The effect of inhaled corticosteroids on Chlamydophila pneumoniae and Mycoplasma pneumoniae infection in children with bronchial asthma. J Infect Chemother. 2009; 15:99–103. PMID: 19396519.
13. Jung CL, Lee MA, Chung WS. Clinical evaluation of the multiplex PCR assay for the detection of bacterial pathogens in respiratory specimens from patients with pneumonia. Korean J Clin Microbiol. 2010; 13:40–46.

14. Hong KB, Choi EH, Lee HJ, Lee SY, Cho EY, Choi JH, et al. Macrolide resistance of Mycoplasma pneumoniae, South Korea, 2000-2011. Emerg Infect Dis. 2013; 19:1281–1284. PMID: 23876792.
15. Cao B, Zhao CJ, Yin YD, Zhao F, Song SF, Bai L, et al. High prevalence of macrolide resistance in Mycoplasma pneumoniae isolates from adult and adolescent patients with respiratory tract infection in China. Clin Infect Dis. 2010; 51:189–194. PMID: 20540621.
16. Isozumi R, Yoshimine H, Morozumi M, Ubukata K, Ariyoshi K. Adult community-acquired pneumonia caused by macrolide resistant Mycoplasma pneumoniae. Respirology. 2009; 14:1206–1208. PMID: 19732389.
17. Oh CE, Choi EH, Lee HJ. Detection of genetic mutations associated with macrolide resistance of Mycoplasma pneumoniae. Korean J Pediatr. 2010; 53:178–183.
Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download