Abstract
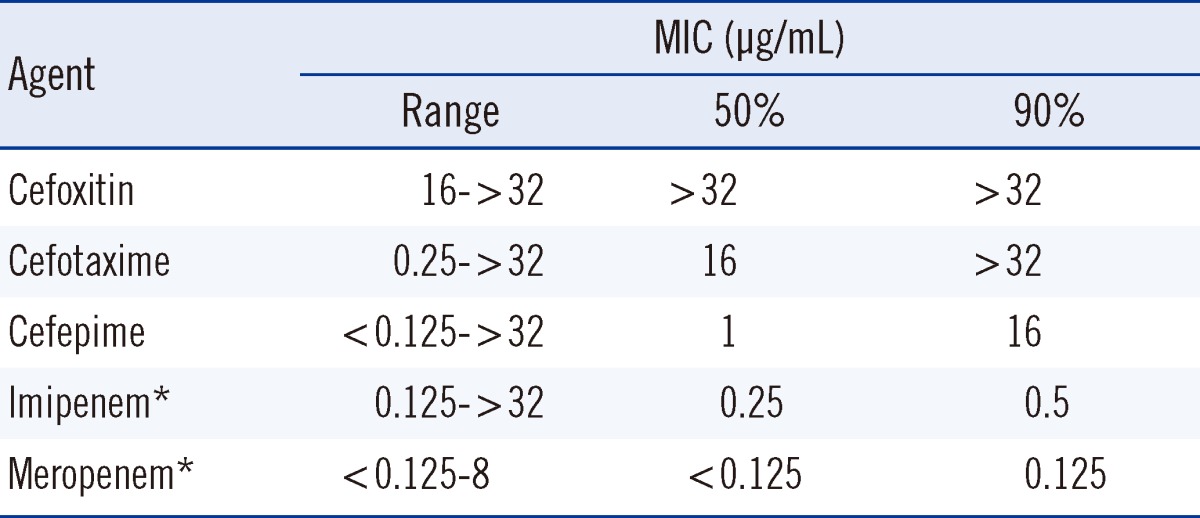
We investigated the occurrence and genetic basis of AmpC β-lactamase (AmpC)-mediated antibiotic resistance, by examining Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a university hospital, from 2007 to 2010. The ampC genes were detected by multiplex AmpC PCR, and AmpC-positive strains were subjected to DNA sequencing. Extended-spectrum β-lactamase (ESBL) production was assessed using the ESBL disk test based on the utilization of boronic acid. Carbapenem-resistant isolates were further investigated by the modified Hodge test, a carbapenemase inhibition test and SDS-PAGE experiments. AmpC expression was detected in 1.6% of E. coli (39 DHA-1, 45 CMY-2, and 1 CMY-1) isolates, 7.2% of K. pneumoniae (39 DHA-1, 45 CMY-2, and 1 CMY-1) isolates, and 2.5% of P. mirabilis (8 CMY-2 and 1 CMY-1) isolates. Of the 198 acquired AmpC producers, 58 isolates (29.3%) also produced an ESBL enzyme. Among the acquired AmpC-producing K. pneumoniae isolates, the minimum inhibitory concentration (MIC) MIC50/MIC90 values for cefoxitin, cefotaxime, cefepime, imipenem, and meropenem were >32/>32, 16/>32, 1/16, 0.25/0.5, and <0.125/0.125 µg/mL, respectively. The MIC values for carbapenem were ≥2 µg/mL for 2 K. pneumoniae isolates, both of which carried the blaDHA-1 gene with a loss of OmpK36 expression, but were negative for carbapenemase production. The acquisition of AmpC-mediated resistance in K. pneumoniae isolates increased, as did the proportion of AmpC and ESBL co-producers among the hospital isolates. The accurate identification of isolates producing AmpCs and ESBLs may aid in infection control and will assist physicians in selecting an appropriate antibiotic regimen.
Unlike other β-lactamases, AmpC β-lactamases (AmpCs) are not inhibited by β-lactamase inhibitors and cephamycins. AmpC-producing organisms are generally resistant to broad-spectrum penicillins, oxyimino- and 7-α-methoxy-cephalosporins, and aztreonam, but are susceptible to cefepime, cefpirome, and carbapenems [1]. Although AmpC acquisition was first detected in Korea in 1988 [2], there remains a lack of awareness about its importance. One reason for this is that most clinical laboratories do not attempt to detect and report this resistance mechanism. Previously, we reported an increased trend in the acquisition of AmpC-mediated resistance in K. pneumoniae isolates in a hospital in Korea, from 2002 to 2004 [3]. In this study, we investigated the occurrence and genetic basis of AmpC-mediated antibiotic resistance by examining Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at the same hospital, from 2007 to 2010.
Between 2007 and 2010, we collected a total of 6,932 consecutive, non-duplicate E. coli (n=5,152), K. pneumoniae (n=1,438), and P. mirabilis (n=367) isolates. Microbial identification was carried out using the Vitek 2 system (bioMerieux Vitek, Hazelwood, MO, USA). Cefoxitin nonsusceptibility (minimum inhibitory concentration [MIC] ≥16 µg/mL) was used as an indicator for AmpC acquisition. The cefoxitin-nonsusceptible isolates were tested by multiplex AmpC PCR [4]. AmpC detection by PCR was further confirmed by ampC gene sequencing. Sequence alignment and analysis was performed on-line using the National Center for Biotechnology Information BLAST algorithm. AmpC PCR-positive isolates were further investigated for β-lactamase production, as well as susceptibility to various antimicrobial agents. Susceptibility was determined by the broth microdilution test, according to the guidelines prescribed by CLSI [5]. The antimicrobial agents used were cefoxitin (Sigma-Aldrich Co., Steinheim, Germany), cefotaxime (Sigma-Aldrich Co.), cefepime (Boryung Co., Ltd., Seoul, Korea), imipenem (Merck & Co., West Point, PA, USA), and meropenem (Yuhan Co., Ltd., Seoul, Korea). AmpC PCR-positive isolates were also analyzed by the boronic acid (BA) disk test [6]. Following the combination disk method (CLSI) extended-spectrum β-lactamase (ESBL) confirmatory methodology, an increase in zone diameter by≥5 mm of CAZ-CA and/or CTX-CA in combination with BA (CAZ-CA-BA and/or CTX-CA-BA) over that of CAZ and/or CTX-containing BA (CAZ-BA and/or CTX-BA) was considered a positive test result for ESBL. Carbapenem-nonsusceptible isolates were further investigated by the modified Hodge test [7] and the carbapenemase inhibition test [8] for carbapenemase production, and were additionally investigated by SDS-PAGE for alterations in outer membrane protein expression [9].
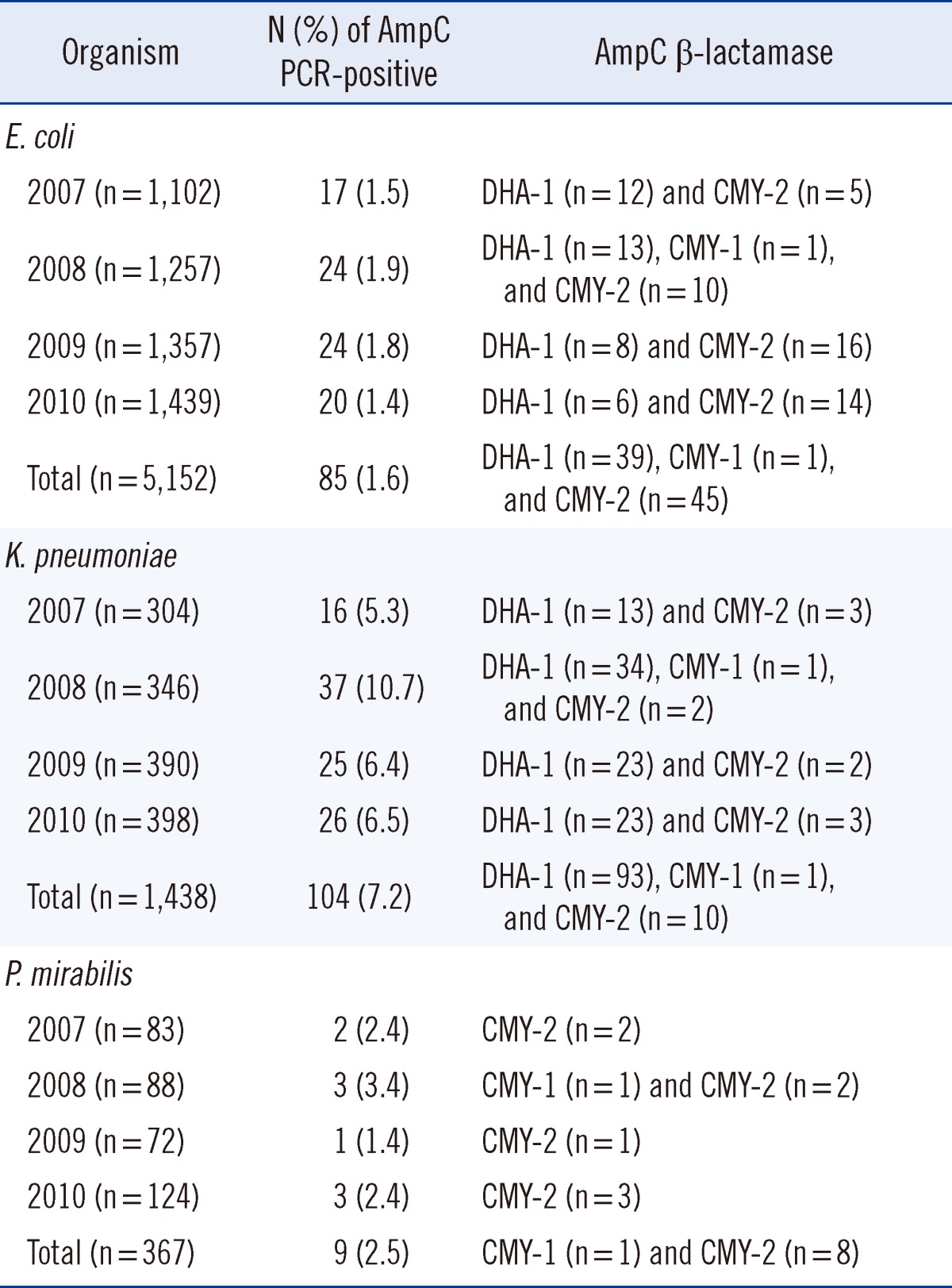
AmpCs (DHA-1, CMY-2, and CMY-1) were detected in 1.6% (85/5,152; 39 DHA-1, 45 CMY-2, and 1 CMY-1) of E. coli isolates, 7.2% (93/1,438; 93 DHA-1, 10 CMY-2, and 1 CMY-1) of K. pneumoniae isolates, and 2.5% (9/367; 8 CMY-2 and 1 CMY-1) of P. mirabilis isolates (Table 1). In Korea, the isolation rate of AmpC-producing E. coli was 1.5% in 2002 [10]. Our previous study revealed that the isolation rate of AmpC-producing K. pneumoniae from 2002 to 2004 was 2.9%, and no AmpC-producing P. mirabilis strains were isolated during that period [3]. While the distribution of AmpC genotypes is similar to that obtained in the previous study [3], of the 198 acquired AmpC producers, 58 isolates (29.3%) also produced an ESBL (data not shown). This rate is much higher than that (8.7%) observed in our previous study [3]. In the case of the acquired AmpC-producing K. pneumoniae isolates, 50/MIC90 values for cefoxitin, cefotaxime, cefepime, imipenem, and meropenem were >32/>32, 16/>32, 1/16, 0.25/0.5, and <0.125/0.125 µg/mL, respectively (Table 2). MIC values for carbapenem were≥2 µg/mL, for only 2 of the K. pneumoniae isolates. Both isolates carried the blaDHA-1 gene with a loss of OmpK36 expression, but were negative for carbapenemase production (data not shown). In Korea, AmpC (DHA-1) expression, in combination with a loss in OmpK35/36 expression contributes to the carbapenem resistance observed in Enterobacteriaceae, and this phenotype is associated with a poor clinical outcome [11, 12].
In summary, there has been an increase in the clinical isolation of acquired AmpC-producing K. pneumoniae, as well as an increase in the isolation of AmpC and ESBL co-producers in recent years in Korea. DHA-1 was the predominant AmpC enzyme in the K. pneumoniae isolates, while CMY-2 was predominantly found in E. coli and P. mirabilis isolates. The accurate identification of isolates producing AmpCs and ESBLs may aid in infection control, and will provide valuable guidance while selecting an appropriate antibiotic regimen.
References
1. Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC β-lactamases. Antimicrob Agents Chemother. 2002; 46:1–11. PMID: 11751104.
2. Bauernfeind A, Chong Y, Schweighart S. Extended broad spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection. 1989; 17:316–321. PMID: 2689349.
3. Song W, Kim JS, Kim HS, Yong D, Jeong SH, Park MJ, et al. Increasing trend in the prevalence of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal ampC gene at a Korean university hospital from 2002 to 2004. Diagn Microbiol Infect Dis. 2006; 55:219–224. PMID: 16545935.
4. Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002; 40:2153–2162. PMID: 12037080.

5. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement, M100-S21. Wayne, PA: CLSI;2011.
6. Song W, Jeong SH, Kim HS, Kim HS, Shin DH, Roh KH, et al. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC β-lactamases and extended-spectrum β-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn Microbiol Infect Dis. 2007; 57:315–318. PMID: 17174510.
7. Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007; 45:2723–2725. PMID: 17581941.
8. Giske CG, Gezelius L, Samuelsen Ø, Warner M, Sundsfjord A, Woodford N. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect. 2011; 17:552–556. PMID: 20597925.
9. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother. 2006; 50:3396–3406. PMID: 17005822.
10. Song W, Kim JS, Kim MN, Kim EC, Park YJ, Yong D, et al. Occurrence and genotypic distributions of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Korea. Korean J Lab Med. 2002; 22:410–416.
11. Song W, Suh B, Choi JY, Jeong SH, Jeon EH, Lee YK, et al. In vivo selection of carbapenem-resistant Klebsiellae pneumonia by OmpK36 loss during meropenem treatment. Diagn Microbiol Infect Dis. 2009; 65:447–449. PMID: 19766430.
12. Shin SY, Bae IK, Kim J, Jeong SH, Yong D, Kim JM, et al. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol. 2012; 61:239–245. PMID: 21940650.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download