Abstract
We used HPLC and AdvanSure real-time PCR (LG Life Sciences, Korea) to retrospectively analyze non-tuberculous mycobacteria (NTM) in 133 clinical specimens. The specimens were culture-positive for NTM and the HPLC method identified 130 strains of mycobacteria from the cultures (97.7%) at the species level. Among the isolates, 48 Mycobacterium. kansasii (36.1%), 39 M. intracellulare (29.3%), 17 M. avium (12.8%), 16 M. abscessus (12.0%), 6 M. fortuitum (4.5%), 2 M. szulgai (1.5%), 2 M. gordonae (1.5%), and 3 unclassified NTM strains (2.3%) were identified. The real-time PCR assay identified 60 NTM-positive specimens (45.1%), 65 negative specimens (48.9%), and 8 M. tuberculosis (TB)-positive specimens (6.0%). The real-time PCR assay is advantageous because of its rapid identification of NTM. However, in our study, the real-time PCR assay showed relatively low sensitivity (45.1%) when using direct specimens including sputum and bronchoalveolar lavage (BAL) fluid. HPLC is useful as it discriminates NTM at the species level, although it is time-consuming and requires specific equipment and technical expertise. A combination of both methods will be helpful for the rapid and accurate identification of mycobacteria in clinical laboratories.
The incidence of non-tuberculous mycobacteria (NTM) infection has increased in the Korean population, with a simultaneous increase in the elderly and immunocompromised populations [1-3]. However, the widespread nature of NTM infection can make it difficult to diagnose [4, 5]. To diagnose mycobacterial infection, laboratories generally adopt biochemical methods, real-time PCR assays, and HPLC. In particular, real-time PCR has several advantages, such as a short turn-around time by the use of direct specimens and high sensitivity and specificity [5, 6]. However, the risk of contamination is high, and therefore, multiple steps are required. In addition, it is difficult to identify NTM at the species level [7, 8]. The AdvanSure TB/NTM real-time PCR assay (LG Life Sciences, Seoul, Korea) cannot discriminate the NTM at the species level; however, it can identify Mycobacterium tuberculosis (TB) and distinguish it from NTM before the growth of acid-fast bacilli (AFB). HPLC is another method used to identify mycobacterial infection as it analyzes the mycolic acids in an organism, and can evaluate various mycobacterial species [9-11]. In this study, we discuss the usefulness of real-time PCR and the HPLC method for isolating NTM.
We retrospectively analyzed the data from 133 NTM positive specimens, including sputum and bronchoalveolar lavage (BAL) fluid, obtained from the department of laboratory medicine, Ulsan University Hospital, University of Ulsan College of Medicine, Ulsan, Korea, between January and December 2011. The specimens were processed for AFB culture and were inoculated in 3% Ogawa solid egg-based medium (Asan Pharmaceutical, Seoul, Korea) and Mycobacteria Growth Indicator Tube liquid medium (Becton Dickinson, Sparks, MD, USA) at the same time, according to the CLSI protocols [12]. The DNA from the 133 clinical specimens was extracted and amplified using the AdvanSure TB/NTM real-time PCR Kit following the manufacturer's protocol. The SLAN real-time PCR detection system (LG Life Science, Seoul, Korea) was used to measure the fluorescence during PCR. A positive result was indicated when the signal was observed in each channel and the cycle threshold (CT) value was less than 35 cycles.
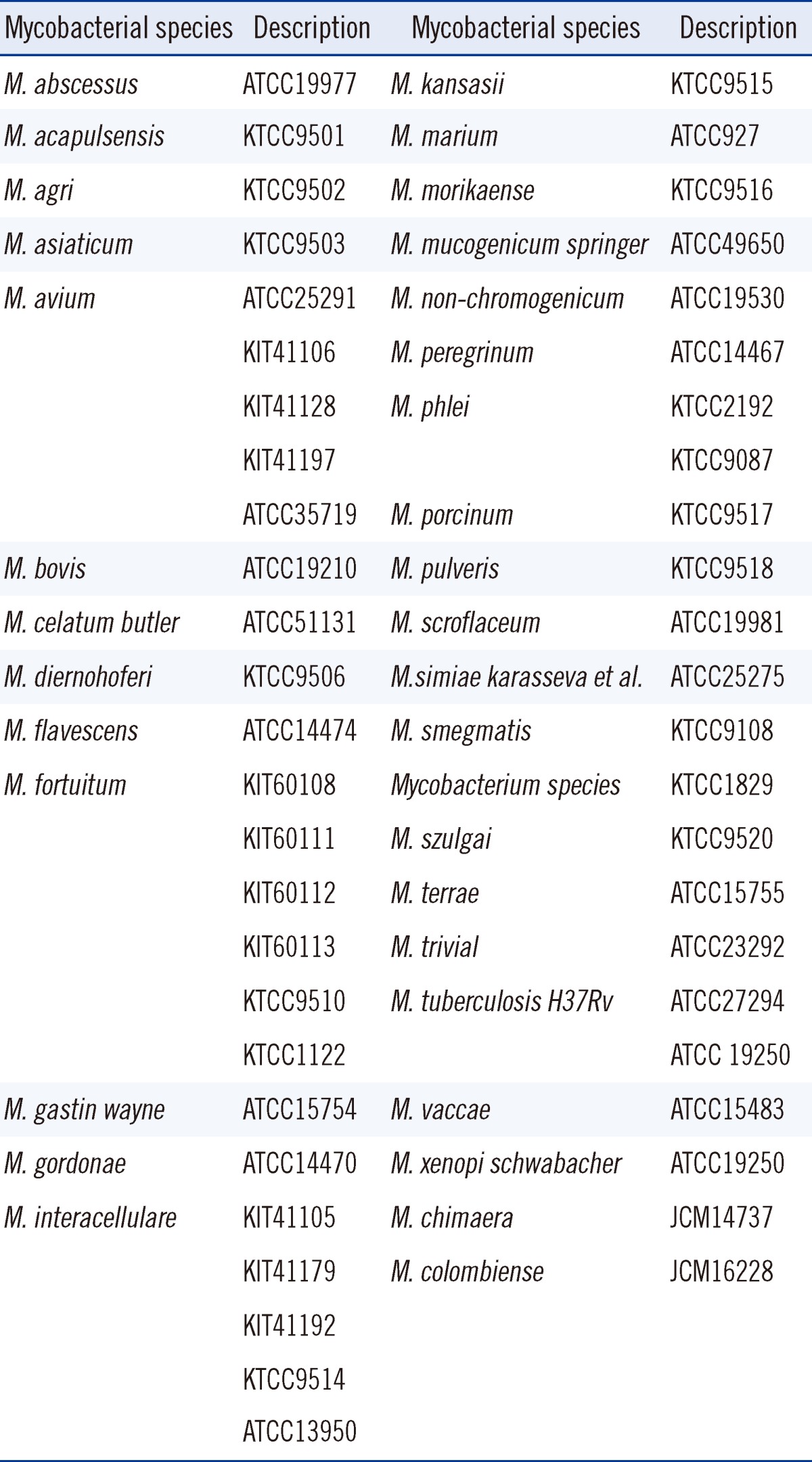
To identify NTM, HPLC was performed, and the results were compared against the patterns of the standard Mycobacterium species [5, 6], which were obtained from the ATCC standard Mycobacterium species, the Korean Type Culture Collection (KTCC) standard Mycobacterium species, the Korean Institute of Tuberculosis (KIT) standard Mycobacterium species, and the Japan Collection of Microorganisms (JCM) standard Mycobacterium species (Table 1). The HPLC instruments were equipped with the Waters 2690 separation module (Milford, MA, USA), a reverse phase analytical cartridge column, (3.9×75 mm), packed with 3 µm silica (Nova-pak C18), and a photodiode array detector (Waters Corporation, Milford, MA, USA). For the low-molecular and high-molecular weight internal standards, 6,7-dimethoxy-4-coumarinylmethyl ester and p-bromophenyl ester were used. The HPLC samples were prepared by cell harvesting, saponification, extraction, and a clarification to isolate mycolic acids. The prepared samples were subjected to HPLC analysis using the ultraviolet detector. Each cluster group was identified by its number of peaks, retention times, and relative peak heights [10].
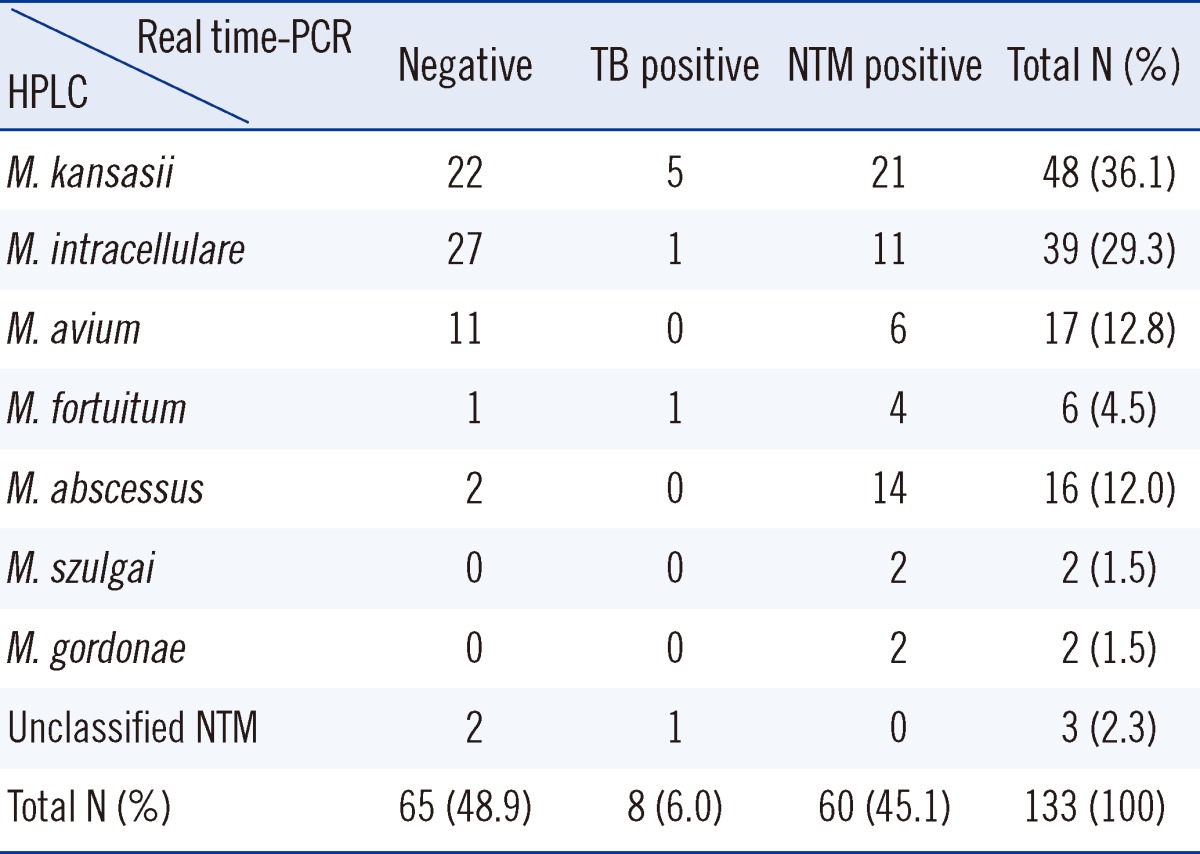
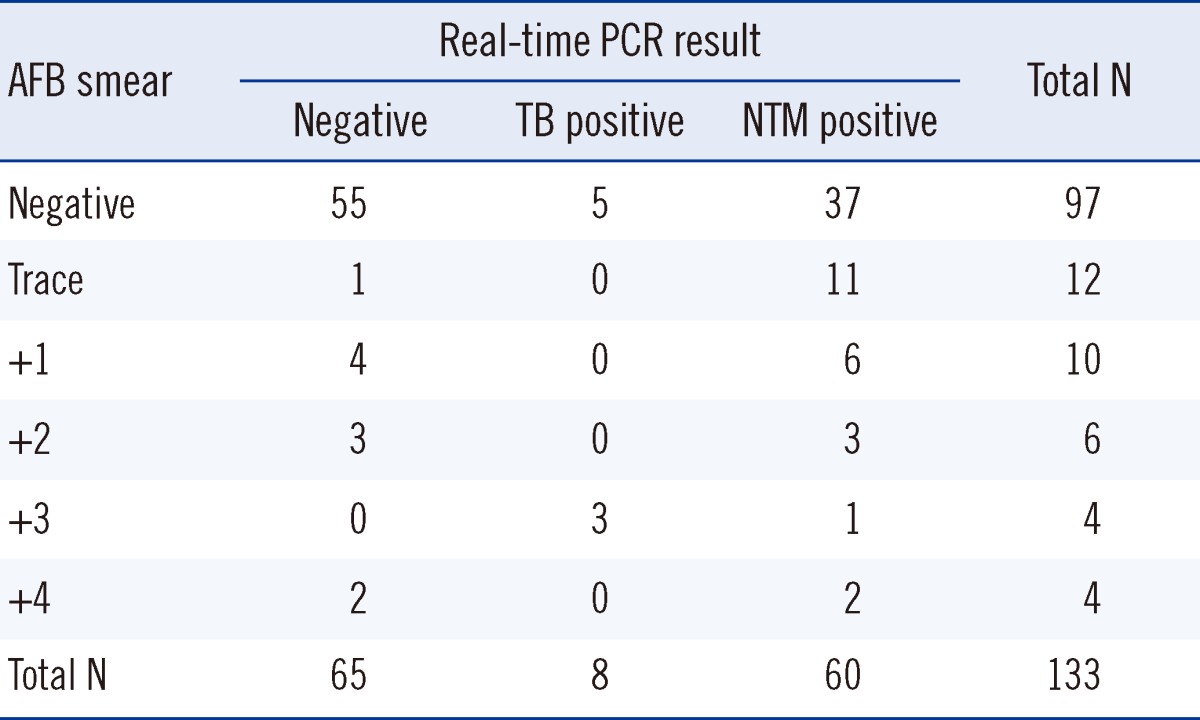
Among the 133 specimens, 60 samples were NTM positive (45.1%), 65 samples were negative (48.9%), and 8 samples were TB positive (6.0%; Table 2) by real-time PCR. Among the 36 specimens that were AFB smear-positive, 23 (63.8%) were NTM-positive according to the real-time PCR assay (Table 3).
The HPLC patterns for the vast majority (97.7%) of the 133 strains of mycobacteria obtained from culture matched those of the 34 standard Mycobacterium species (Table 1). Three specimens (2.3%) did not match any of the standards and were grouped into the "unclassified NTM" category (Table 2). In total, 7 NTM species were identified and the rest were termed as unclassified NTM. In the single-cluster group, 3 species, namely, M. kansasii, M. szulgai and M. gordonae, were identified. In the double-cluster group, there were 4 NTM species, M. intracellulare, M. abscessus, M. avium, and M. fortuitum. The distribution of identified NTM species was as follows: 48 M. kansasii (36.1%), 39 M. intracellulare (29.3%), 17 M. avium (12.8%), 16 M. abscessus (12.0%), 6 M. fortuitum (4.5%), 2 M. szulgai (1.5%), 2 M. gordonae (1.5%), and 3 unclassified NTM (2.3%; Table 2).
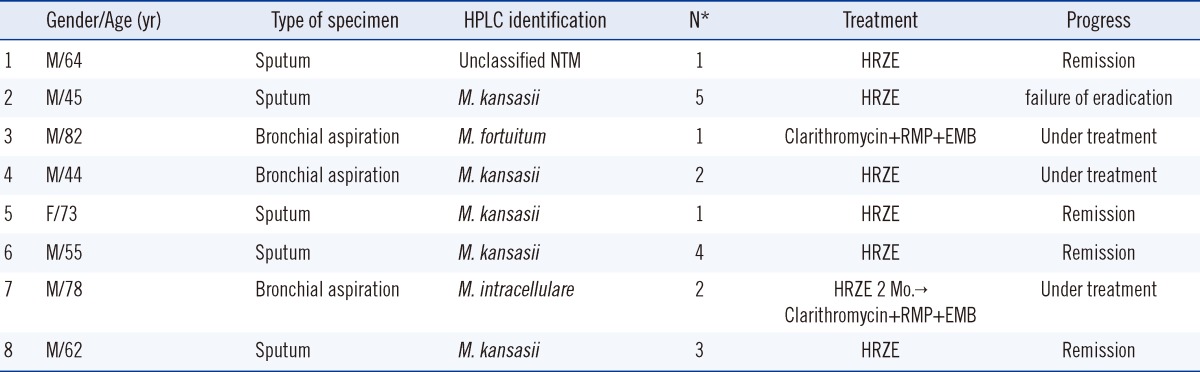
Discrepant results were obtained for 8 specimens that were positive for TB by real-time PCR, but positive for NTM by HPLC (Table 4). We performed the real-time PCR assay on the culture isolates from the 8 discrepant samples, and found 7 specimens positive for NTM and only the first specimen contained both TB and NTM strains. Species analysis was limited because we did not use a more precise method such as DNA sequencing.
The Centers for Disease Control and Prevention (CDC) suggested an algorithm for the diagnosis of mycobacterial infection that includes AFB staining, AFB culture, nucleic acid amplification testing, and HPLC [13]. The real-time PCR assay is useful as it has the advantage of rapid identification of mycobacteria by using direct specimens. This test could provide information regarding the type of mycobacteria present, which could conceivably aid in the selection of subsequent tests and may decrease diagnostic delays by reducing unnecessary testing. The AdvanSure real-time PCR assay has an overall sensitivity of 66.7-100%. Specifically, 86.8-100% specificity was obtained using AFB smear-positive specimens [6, 14, 15], and 55.6-75% sensitivity in NTM isolation assays [6, 15]. In our study, the real-time PCR assay showed 45.1% sensitivity (95% confidence interval, 36.5-53.9%) among the clinical specimens determined to be NTM-culture-positive. Among the AFB smear-positive specimens, the real-time PCR assays showed 63.8% sensitivity (95% confidence interval, 46.2-78.6%) with respect to NTM detection. This difference is attributed to the numbers involved in the study and the high proportion of smear-negative results among the samples (97/133, 72.9%). Real-time PCR is limited by false-positive and false-negative results that dictate the need to perform microbial cultures to confirm the findings.
On the other hand, HPLC assay discriminates the NTM at the species level (97.7%), although it requires particular equipment and technical expertise. With respect to the species distribution, M. kansasii, M. intracellulare, M. abscessus, and M. avium were the most common. Comparatively, Koh et al. [16], reported that M. avium and M. abscessus were most often found, which may reflect the local characteristics.
In conclusion, real-time PCR is useful allowing the rapid identification of mycobacteria by direct specimens; however, it has relatively low sensitivity. The HPLC method has high sensitivity, and is also useful for determining the species level identify of organisms, although the culture growth requirement is time-consuming. A combination of both methods will enable the rapid and accurate identification of mycobacteria in clinical laboratories.
References
1. Subcommittee of the Joint Tuberculosis Committee of the British Thoracic Society. Management of opportunist mycobacterial infections: Joint Tuberculosis Committee Guidelines 1999. Thorax. 2000; 55:210–218. PMID: 10679540.
2. Daley CL. Nontuberculous mycobacterial disease in transplant recipients: early diagnosis and treatment. Curr Opin Organ Transplant. 2009; 14:619–624. PMID: 19745733.

3. Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003; 163:1009–1021. PMID: 12742798.
4. Falkinham JO 3rd. Nontuberculous mycobacteria in the environment. Clin Chest Med. 2002; 23:529–551. PMID: 12370991.

5. Wang HY, Jin H, Bang H, Choi YI, Park EM, Koh WJ, et al. Evaluation of MolecuTech real MTB-ID for MTB/NTM detection using direct specimens. Korean J Clin Microbiol. 2011; 14:103–109.

6. Hwang S, Oh KJ, Jang IH, Uh Y, Yoon KJ, Kim HY, et al. Evaluation of the diagnostic performance of the AdvanSure TB/NTM real-time PCR kit for detection of mycobacteria. Korean J Clin Microbiol. 2011; 14:55–59.

7. Park CM, Heo SR, Park KU, Song J, Lee JH, Lee CT, et al. Isolation of nontuberculous mycobacteria using polymerase chain reaction-restriction fragment length polymorphism. Korean J Lab Med. 2006; 26:161–167. PMID: 18156719.

8. Burman WJ, Reves RR. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis. 2000; 31:1390–1395. PMID: 11096008.
9. Jeong J, Kim SR, Chang CL, Lee SH. Identification of mycobacteria species by HPLC and species distribution during five years at Ulsan university hospital. Korean J Lab Med. 2008; 28:24–33. PMID: 18309252.

10. Butler WR, Guthertz LS. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin Microbiol Rev. 2001; 14:704–726. PMID: 11585782.
11. Jeong J, Kim SR, Lee SH, Lim JH, Choi JI, Park JS, et al. The use of high performance liquid chromatography to speciate and characterize the epidemiology of mycobacteria. Lab Med. 2011; 42:612–617.

12. Clinical and Laboratory Standards Institute. Laboratory detection and identification of mycobacteria; approved guideline M48-A. Wayne, PA: CLSI;2008.
13. Shinnick TM, Iademarco MF, Ridderhof JC. National plan for reliable tuberculosis laboratory services using a systems approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR Recomm Rep. 2005; 54:1–12. PMID: 15829862.
14. Chang HE, Heo SR, Yoo KC, Song SH, Kim SH, Kim HB, et al. Detection of mycobacterium tuberculosis complex using real-time polymerase chain reaction. Korean J Lab Med. 2008; 28:103–108. PMID: 18458505.

15. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. Evaluation of the performances of AdvanSure TB/NTM real time PCR kit for detection of mycobacteria in respiratory specimens. Korean J Lab Med. 2008; 28:34–38. PMID: 18309253.

16. Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006; 129:341–348. PMID: 16478850.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download