Abstract
Background
Active screening for vancomycin-resistant enterococci (VRE) using rectal specimens is recommended to limit the spread of antimicrobial resistance within certain high-risk populations. We evaluated the diagnostic performance of Vancomycin Resistance 3 Multiplexed Tandem PCR assay (AusDiagnostics, Australia), a rapid multiplex real-time PCR assay that detects vanA and/or vanB.
Methods
Two-hundred-and-eleven rectal swabs from Hematology and Oncology unit were submitted for VRE surveillance via direct detection of vanA and/or vanB by culture and by using Vancomycin Resistance 3 Multiplexed Tandem PCR assay. Enterococci were identified to the species level by using standard biochemical tests and BD Phoenix Automated Microbiology System (BD Diagnostic Systems, USA). Vancomycin susceptibility of enterococci was determined using Etest (BioMerieux, France).
Results
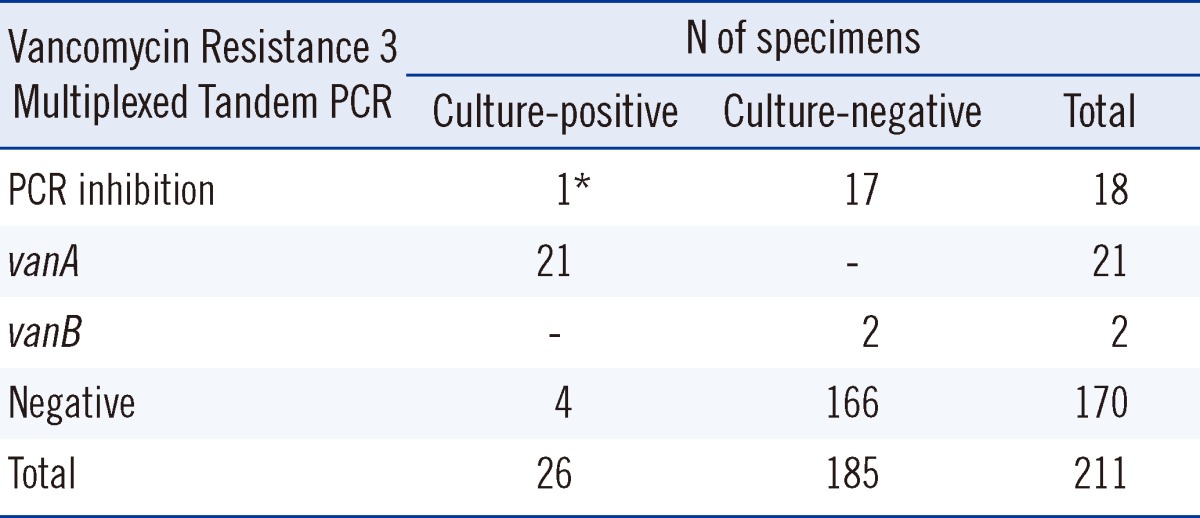
Compared to the culture method, Vancomycin Resistance 3 Multiplexed Tandem PCR assay had a sensitivity of 84.0%, specificity of 98.8%, positive predictive value (PPV) of 91.3%, and negative predictive value (NPV) of 97.6%. The assay failed to detect 18 (8.5%) specimens because of the presence of PCR inhibitors; of the remaining 193 specimens, 25 (12.9%) were positive, 23 for vanA, and 2 for vanB. Although both sensitivity and specificity for vanA VRE was 100% compared to the culture method, all vanB-positive specimens tested negative by VRE culture.
Conclusions
Vancomycin Resistance 3 Multiplexed Tandem PCR assay is a rapid and laborsaving option for VRE surveillance for direct use on rectal swabs. However, the high rate of PCR failure owing to the inhibitors in the specimens and the low specificity for vanB should be considered when interpreting the results.
The emerging resistance to glycopeptide antibiotics (vancomycin and teicoplanin) observed in enterococci is soon becoming one of the most serious issues related to infection control. The first outbreak of vancomycin-resistant Enterococcus (VRE) in Turkey was reported in 1998 [1], and the increase in its prevalence since then has been associated with higher healthcare costs and mortality rates [2]. Vancomycin resistance in enterococci is mainly due to the acquisition of vanA and vanB genes, which have been primarily detected in Enterococcus faecium [3].
Asymptomatic intestinal colonization with VRE is widely reported, and it can act as a reservoir for dissemination and subsequent infection [4-6]. Effective infection control and prevention measures can reduce the colonization and transmission rates, thus reducing the infection rate. Early diagnosis of VRE colonization is, therefore, critical to reduce the incidence of VRE infections and outbreaks. Culture-based methods are typically used for the detection of VRE, which requires 24-72 hr for isolation, identification, and susceptibility testing [7, 8]. However, a screening assay that could detect VRE colonization in < 24 hr would prevent the spread of VRE by allowing earlier implementation of appropriate barrier precautions. Several nucleic acid amplification tests have been developed and evaluated for the detection of VRE, but quite a few of these require complex regimens for extraction and detection [9-12] or an enrichment step involving the use of a selective enrichment broth [13, 14] or isolates recovered from solid medium [15, 16]. The Vancomycin Resistance 3 Multiplexed Tandem PCR kit (AusDiagnostics, Alexandria, Australia) is designed for direct use on rectal swabs for active VRE surveillance. In this study, we aimed to evaluate this kit for early detection of VRE colonization.
A total of 211 non-duplicate rectal swabs collected at the Hematology and Oncology unit at Akdeniz University Faculty of Medicine during an outbreak and submitted to the Clinical Microbiology laboratory were used in this study. This study was performed in April 2012 in accordance with the institutional VRE surveillance program.
Two rectal swab specimens were collected from all patients, and one was inoculated into Enterococcosel broth containing 6 µg/mL vancomycin (BD Diagnostic Systems, Sparks, MD, USA) and incubated in 5-10% CO2 at 35℃ for 24-72 hr. Black discoloration or cloudiness in the broth was considered positive; the culture was then subcultured on Enterococcosel agar containing 6 µg/mL vancomycin (BD Diagnostic Systems). Cultures were considered negative, if no growth was observed on the third day. Black colonies on Enterococcosel agar were identified as potential VREvancomycin-resistant enterococci; these were then subcultured to sheep blood agar plates and incubated at 35℃ for 24 hr. Catalase-negative, gram-positive cocci positive for leucine aminopeptidase (LAP; Remel, Lenexa, KS, USA) and L-pyrolidonyl-β-naphthylamide (PYR; Remel) were further identified using colony morphology, methyl-α-D-glucopyranoside (MDG; Sigma, Taufkirchen, Germany) test, and motility. Species identification and antimicrobial susceptibility testing was performed by using BD Phoenix System (BD Diagnostic Systems). Enterococcus faecalis strain (ATCC 29212) was used as a the control strain in the identification assays. The minimum inhibitory concentrations (MICs) of vancomycin and teicoplanin were determined by the E-test method according to the manufacturer's recommendations. The van gene was typed using the BD GeneOhm™ VanR Assay (BD Diagnostic Systems).
All the specimens were studied with the PCR assay according to the manufacturer's instructions. Vancomycin Resistance 3 Multiplex Tandem PCR assay was configured to screen for VRE colonization in hospital patients by testing perianal and/or rectal swabs for the presence of vanA and vanB genes. The assay uses the principle of Multiplexed Tandem PCR employing 2 sequential PCR steps. Step 1 is multiplex amplification using primers homologous to all targets in the panel. The product from Step 1 is then diluted into individual wells for real-time PCR (Step 2) using primers "nested inside" the primers used for Step 1. This process is automated by the Easy-Plex system (AusDiagnostics). The Rotor-Gene Q thermal cycler (Qiagen, Hilden, Germany) was used for DNA amplification, which was measured by the increase in fluorescence when Eva-Green™ dye is incorporated into the DNA being formed. The 3 targets (vanA, vanB, and an internal control) were amplified together in Step 1 by using 3 tube strips for 24 samples. Step 2 is performed in individual wells fused together into a 72-position Gene-Disc. The assay could be completed in approximately 90 min for every 24 samples.
Of the 211 rectal swab samples, 18 (8.5%) were not effectively amplified by the PCR-presumably because of the presence of PCR inhibitors in the samples; one of them tested positive for VRE by using the enrichment culture method. Samples showing PCR inhibition were excluded from the study, leaving 193 samples for consideration. Comparative results for the culture-based method and PCR are listed in Table 1 and described in detail in the following results section. The PCR assay was assessed using the results obtained with the enrichment culture method as the gold standard. Of the 25 positive PCR results, 21 were positive for vanA, and 2, for vanB. All the 21 vanA-positive results tested positive for VRE by culture method; we confirmed vanA genotype by using BD GeneOhm™ VanR Assay. None of the vanB-positive PCR results confirmed with those obtained with the culture method. Four of the samples that tested positive in the culture method tested negative in the PCR assay. Compared to the culture method, the Vancomycin Resistance 3 Multiplexed Tandem PCR assay yielded a sensitivity of 84.0%, specificity of 98.8%, positive predictive value (PPV) of 91.3%, and negative predictive value (NPV) of 97.6%.
The increasing global prevalence of VRE [17-22] has led to increased interest in screening of patients for colonization and in development of methods for rapid, sensitive, and reliable detection of VRE [23]. Various commercial phenotypic and genotypic assays with different sensitivity and specificity are available for VRE screening [12, 24-30]; however, genotypic assays are generally more rapid and sensitive [28, 31, 32]. The Cepheid GeneXpert vanA/vanB assay, BD GeneOhm VanR assay, and other commercially available assays have high sensitivity and specificity for detecting vanA-positive specimens [24, 28-30], but a low specificity due to the comparably high rates of apparent false-positive vanB-positive specimens [12, 28]. The low specificity of detection of the vanB gene by various assays has been explained by the presence of commensal bacteria of the fecal flora carrying the vanB gene [8, 12, 13, 25, 28, 33]. Consistent with these findings, the 2 vanB genes we detected by PCR were not confirmed using the culture method. To the best of our knowledge, this is the first study involving Vancomycin Resistance 3 Multiplexed Tandem PCR assay, and like all other methods studied for the active surveillance and detection of VRE, it produced false-positive results due to vanB. Therefore, it has been suggested that follow-up culture should be performed on all vanB-positive specimens [12, 27]. The vanB genotype has never been detected in this hospital until date [1, 34]. In our sample set, vanB was present in 8.0% (2/25) of the total number of specimens, and in this setting, confirmation with the culture method has prevented unnecessary precautions during isolation of strains.
In our study, 18 (8.5%) of the 211 rectal swab samples contained PCR inhibitors. Because one of these samples tested positive for VRE using the culture method, this finding highlights a disadvantage of the assay. PCR inhibitors are often present in stool samples, and may originate from dietary components, polysaccharides, or chlorophyll from herbs and vegetables, bile salts, urea, glycolipids, hemoglobin, and heparin [35]. Although automated nucleic acid extraction systems improve the consistency and throughput of PCR tests, these systems may prove insufficient for removal of PCR inhibitors [36]. Although various protocols have been developed to remove PCR inhibitors (e.g., heat treatment before PCR, chloroform extraction, treatment with activated carbon, sample dilution), they may affect the sensitivity of the assay or lead to false-negative results [35].
In our study, 12.9% of the specimens tested positive for VRE, and all the strains were found to display vanA phenotype. This is not an unexpected result, considering that there has been only 1 report describing the isolation of vanB-positive E. faecium in Turkey [37]. As a part of our study, we collected samples during an outbreak period from the Hematology and Oncology unit and from pediatric and adult patients. Many of them could have been treated with antibiotics, which might increase the selective pressure for VRE. However, as the antibiotic therapy received by the patients was not documented for the present study, the effects of antibiotics could not be compared.
The Vancomycin Resistance 3 Multiplexed Tandem PCR kit had an excellent NPV and PPV for the detection of vanA. Because the kit can rapidly identify patients not carrying vancomycin-resistance genes and those who have acquired the vanA and vanB genes, healthcare professionals can, within 3 hr of patient admission, determine appropriate infection control policies to prevent cross infections. Strains testing positive for vanA can be rapidly identified as VRE, but strains testing positive for vanB need to be confirmed by the culture method.
In conclusion, direct application of Vancomycin Resistance 3 Multiplexed Tandem PCR assay on rectal swabs is a reliable option to give rapid and accurate results for vanA-VRE surveillance.
Acknowledgements
The authors would like to thank infection control committee of Akdeniz University Faculty of Medicine.
References
1. Colak D, Naas T, Gunseren F, Fortineau N, Ogunc D, Gultekin M, et al. First outbreak of vancomycin-resistant enterococci in a tertiary hospital in Turkey. J Antimicrob Chemother. 2002; 50:397–401. PMID: 12205065.

2. Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008; 52:297–308. PMID: 18279340.
3. Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006; 42(Suppl 1):S25–S34. PMID: 16323116.

4. Matar MJ, Safdar A, Rolston KV. Relationship of colonization with vancomycin-resistant enterococci and risk of systemic infection in patients with cancer. Clin Infect Dis. 2006; 42:1506–1507. PMID: 16619176.

5. Werner G, Serr A, Schütt S, Schneider C, Klare I, Witte W, et al. Comparison of direct cultivation on a selective solid medium, polymerase chain reaction from an enrichment broth, and the BD GeneOhm™ VanR Assay for identification of vancomycin-resistant enterococci in screening specimens. Diagn Microbiol Infect Dis. 2011; 70:512–521. PMID: 21767707.

6. Olivier CN, Blake RK, Steed LL, Salgado CD. Risk of vancomycin-resistant Enterococcus (VRE) bloodstream infection among patients colonized with VRE. Infect Control Hosp Epidemiol. 2008; 29:404–409. PMID: 18419361.
7. Novicki TJ, Schapiro JM, Ulness BK, Sebeste A, Busse-Johnston L, Swanson KM, et al. Convenient selective differential broth for isolation of vancomycin-resistant enterococcus from fecal material. J Clin Microbiol. 2004; 42:1637–1640. PMID: 15071018.
8. Van Horn KG, Gedris CA, Rodney KM, Mitchell JB. Evaluation of commercial vancomycin agar screen plates for detection of vancomycin-resistant enterococci. J Clin Microbiol. 1996; 34:2042–2044. PMID: 8818911.

9. Domingo MC, Huletsky A, Giroux R, Boissinot K, Picard FJ, Lebel P, et al. High prevalence of glycopeptide resistance genes vanB, vanD, and vanG not associated with enterococci in human fecal flora. Antimicrob Agents Chemother. 2005; 49:4784–4786. PMID: 16251331.
10. Palladino S, Kay ID, Flexman JP, Boehm I, Costa AM, Lambert EJ, et al. Rapid detection of vanA and vanB genes directly from clinical specimens and enrichment broths by real-time multiplex PCR assay. J Clin Microbiol. 2003; 41:2483–2486. PMID: 12791869.
11. Paule SM, Trick WE, Tenover FC, Lankford M, Cunningham S, Stosor V, et al. Comparison of PCR assay to culture for surveillance detection of vancomycin-resistant enterococci. J Clin Microbiol. 2003; 41:4805–4807. PMID: 14532226.

12. Sloan LM, Uhl JR, Vetter EA, Schleck CD, Harmsen WS, Manahan J, et al. Comparison of the Roche LightCycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. J Clin Microbiol. 2004; 42:2636–2643. PMID: 15184445.
13. Ballard SA, Grabsch EA, Johnson PD, Grayson ML. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob Agents Chemother. 2005; 49:77–81. PMID: 15616278.
14. Drews SJ, Johnson G, Gharabaghi F, Roscoe M, Matlow A, Tellier R, et al. A 24-hour screening protocol for identification of vancomycin-resistant Enterococcus faecium. J Clin Microbiol. 2006; 44:1578–1580. PMID: 16597899.
15. Petrich A, Luinstra K, Page B, Callery S, Stevens D, Gafni A, et al. Effect of routine use of a multiplex PCR for detection of vanA- and vanB-mediated enterococcal resistance on accuracy, costs and earlier reporting. Diagn Microbiol Infect Dis. 2001; 41:215–220. PMID: 11777663.
16. Sahm DF, Free L, Smith C, Eveland M, Mundy LM. Rapid characterization schemes for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 1997; 35:2026–2030. PMID: 9230375.

17. Brown DF, Hope R, Livermore DM, Brick G, Brougthon K, George RC, et al. Non-susceptibility trends among enterococci and non-pneumococcal streptococci from bacteraemias in the UK and Ireland, 2001 to 2006. J Antimicrob Chemother. 2008; 62(Suppl 2):ii75–ii85. PMID: 18819982.
18. Henard S, Lozniewski A, Aissa N, Jouzeau N, Rabaud C. Evaluation of the duration of vanA vancomycin-resistant Enterococcus faecium carriage and clearance during a large-scale outbreak in a region of eastern France. Am J Infect Control. 2011; 39:169–171. PMID: 20971530.
19. Sagel U, Schulte B, Heeg P, Borgmann S. Vancomycin-resistant enterococci outbreak, Germany, and calculation of outbreak start. Emerg Infect Dis. 2008; 14:317–319. PMID: 18258130.

20. Ergani-Ozcan A, Naas T, Baysan BO, Ogunc D, Inan D, Colak D, et al. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J Antimicrob Chemother. 2008; 61:1033–1039. PMID: 18319236.

21. Theilacker C, Jonas D, Huebner J, Bertz H, Kern WV. Outcomes of invasive infection due to vancomycin-resistant Enterococcus faecium during a recent outbreak. Infection. 2009; 37:540–543. PMID: 19669085.

22. Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008; 13:pii: 19046.

23. Vonberg RP, Chaberny IF, Kola A, Mattner F, Borgmann S, Dettenkofer M, et al. Prevention and control of the spread of vancomycin-resistant enterococci: results of a workshop held by the German Society for Hygiene and Microbiology. Anaesthesist. 2007; 56:151–157. PMID: 17171367.
24. Bourdon N, Bérenger R, Lepoultier R, Mouet A, Lesteven C, Borgey F, et al. Rapid detection of vancomycin-resistant enterococci from rectal swabs by the Cepheid Xpert vanA/vanB assay. Diagn Microbiol Infect Dis. 2010; 67:291–293. PMID: 20542208.
25. Grabsch EA, Ghaly-Derias S, Gao W, Howden BP. Comparative study of selective chromogenic (chromID VRE) and bile esculin agars for isolation and identification of vanB-containing vancomycin-resistant enterococci from feces and rectal swabs. J Clin Microbiol. 2008; 46:4034–4036. PMID: 18832121.
26. Ledeboer NA, Das K, Eveland M, Roger-Dalbert C, Mailler S, Chatellier S, et al. Evaluation of a novel chromogenic agar medium for isolation and differentiation of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis isolates. J Clin Microbiol. 2007; 45:1556–1560. PMID: 17329453.
27. Stamper PD, Shulder S, Bekalo P, Manandhar D, Ross TL, Speser S, et al. Evaluation of BBL CHROMagar VanRE for detection of vancomycin-resistant Enterococci in rectal swab specimens. J Clin Microbiol. 2010; 48:4294–4297. PMID: 20739492.

28. Stamper PD, Cai M, Lema C, Eskey K, Carroll KC. Comparison of the BD GeneOhm VanR assay to culture for identification of vancomycin-resistant enterococci in rectal and stool specimens. J Clin Microbiol. 2007; 45:3360–3365. PMID: 17704282.

29. Babady NE, Gilhuley K, Cianciminio-Bordelon D, Tang YW. Performance characteristics of the Cepheid Xpert vanA assay for rapid identification of patients at high risk for carriage of vancomycin-resistant Enterococci. J Clin Microbiol. 2012; 50:3659–3663. PMID: 22972822.
30. Usacheva EA, Ginocchio CC, Morgan M, Maglanoc G, Mehta MS, Tremblay S, et al. Prospective, multicenter evaluation of the BD GeneOhm VanR assay for direct, rapid detection of vancomycin-resistant Enterococcus species in perianal and rectal specimens. Am J Clin Pathol. 2010; 134:219–226. PMID: 20660324.
31. Naas T, Fortineau N, Snanoudj R, Spicq C, Durrbach A, Nordmann P. First nosocomial outbreak of vancomycin-resistant Enterococcus faecium expressing a VanD-like phenotype associated with a vanA genotype. J Clin Microbiol. 2005; 43:3642–3649. PMID: 16081891.
32. Song JH, Ko KS, Suh JY, Oh WS, Kang CI, Chung DR, et al. Clinical implications of vancomycin-resistant Enterococcus faecium (VRE) with VanD phenotype and vanA genotype. J Antimicrob Chemother. 2008; 61:838–844. PMID: 18230690.
33. Graham M, Ballard SA, Grabsch EA, Johnson PD, Grayson ML. High rates of fecal carriage of nonenterococcal vanB in both children and adults. Antimicrob Agents Chemother. 2008; 52:1195–1197. PMID: 18180361.
34. Ongut G, Kilinckaya H, Baysan BO, Ogunc D, Colak D, Inan D, et al. Evaluation of Brilliance VRE agar for the detection of vancomycin-resistant enterococci in rectal swab specimens. J Med Microbiol. 2013; 62:661–662. PMID: 23355311.

35. Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors-occurrence, properties and removal. J Appl Microbiol. 2012; 113:1014–1026. PMID: 22747964.
36. Verheyen J, Kaiser R, Bozic M, Timmen-Wego M, Maier BK, Kessler HH. Extraction of viral nucleic acids: comparison of five automated nucleic acid extraction platforms. J Clin Virol. 2012; 54:255–259. PMID: 22503856.

37. Coşkun FA, Mumcuoğlu I, Aksu N, Karahan ZC, Us E, Tekeli FA, et al. Phenotypic and genotypic traits of vancomycin-resistant enterococci in a public hospital: the first vanB-positive Enterococcus faecium isolates. Mikrobiyol Bul. 2012; 46:276–282. PMID: 22639316.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download