Abstract
Sphingobacterium spiritivorum has been rarely isolated from clinical specimens of immunocompromised patients, and there have been no case reports of S. spiritivorum infection in Korea to our knowledge. We report a case of S. spiritivorum bacteremia in a 68-yr-old woman, who was diagnosed with acute myeloid leukemia and subsequently received chemotherapy. One day after chemotherapy ended, her body temperature increased to 38.3℃. A gram-negative bacillus was isolated in aerobic blood cultures and identified as S. spiritivorum by an automated biochemical system. A 16S rRNA sequencing analysis confirmed that the isolate was S. spiritivorum. The patient received antibiotic therapy for 11 days but died of septic shock. This is the first reported case of human S. spiritivorum infection in Korea. Although human infection is rare, S. spiritivorum can be a fatal opportunistic pathogen in immunocompromised patients.
Sphingobacterium species are non-fermentative, gram-negative rods that are positive for biochemical tests such as catalase and oxidase production, but are negative for that of indole [1]. Sphingobacterium species were previously described as unnamed bacteria (part of Centers for Disease Control and Prevention group IIk). Holmes et al. [2, 3] proposed the genus name Flavobacterium for the bacteria, while in 1983, Yabuuchi et al. [4] first proposed the name Sphingobacterium for the genus. The genus Sphingobacterium was created to classify organisms that contain large amounts of sphingophospholipid compounds in their cell membranes, and have other taxonomic features that distinguish them from Flavobacterium species [4].
Sphingobacterium species have usually been isolated from soil, plants, foodstuffs, and water sources, but the isolation of the species from human clinical specimens has been rarely reported worldwide [5]. Furthermore, Sphingobacterium spiritivorum have been rarely isolated from clinical specimens of immunocompromised patients, and there have been no case reports of S. spiritivorum infection in Korea to our knowledge. We report a case of S. spiritivorum bacteremia in a patient, who had undergone chemotherapy for acute myeloid leukemia.
A 68-yr-old woman was admitted to our hospital for dyspnea that had lasted for 7 days. She had no history of smoking or pulmonary diseases. A physical examination did not find lymphadenopathy or organomegaly. An initial complete blood cell count showed the following results: hemoglobin, 11.3 g/dL; white blood cell counts, 2.86×109/L (neutrophils, 40%; lymphocytes, 57%; and monocytes, 3%); platelet counts, 47×109/L. A peripheral blood smear revealed pancytopenia with no leukemic blasts.
A bone marrow (BM) examination was performed to evaluate pancytopenia. The BM aspirate smears and cytochemical staining showed increased myeloblasts (40%) and other myeloid precursors. The patient was diagnosed with acute myeloid leukemia, not otherwise specified, based on the 2008 WHO classification system [6].
She received chemotherapy of cytarabine 160 mg and idarubicin 20 mg for 3 days, and idarubucin 20 mg for 4 days. One day after the chemotherapy ended, her body temperature increased to 38.3℃. Her blood pressure, pulse rate, and respiratory rate were 90/70 mmHg, 110/min, and 20/min, respectively. Her C-reactive protein (CRP) level increased to 6.31 mg/dL, and her procalcitonin level was 0.08 ng/mL (reference range: <0.05 ng/mL).
Four sets of blood cultures were collected: 2 sets from peripheral veins and 2 sets from the central venous catheter. The patient received empirical antibiotic therapy with intravenous cefepime (2 g every 12 hr). After 1 day of incubation, gram-negative bacilli grew in an aerobic culture bottle that contained the culture from the central venous catheter. After 2 days of incubation, Gram-negative bacilli also grew in an aerobic culture bottle containing the culture from the peripheral vein (Fig. 1A). The positive culture broth was inoculated onto a blood agar plate (BAP) and a MacConkey agar plate (MAC) and incubated for 48 hr at 37℃ with 5% CO2. Non-hemolytic, light yellow-colored colonies grew on the BAP (Fig. 1B). A few colonies also grew on the MAC. Catalase and oxidase tests were positive, but the indole test was negative. The organism was identified as S. spiritivorum with 98.0% probability using the Vitek 2 Gram-Negative Identification card (BioMérieux inc., Marcy-l'Etoile, France).
To confirm the identity of the isolate, 16S ribosomal RNA (rRNA) sequencing analysis was performed. InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA) was used to extract the bacterial genomic DNA, and the first 500 base pairs on the 5' end of the 16S rRNA gene were amplified and sequenced using MicroSeq 500 16S rDNA Bacterial Identification PCR and Sequencing Kits (Applied Biosystems, Foster City, CA, USA). The sequencing product was analyzed on a 3130 Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions.
The resulting sequence of the patient-derived isolate was compared with sequences stored in GenBank (http://www.ncbi.nlm.nih.gov/genbank), EMBL (The European Molecular Biology Laboratory, http://www.ebi.ac.uk/embl), RDP-II (The Ribosomal Database Project, http://rdp.cme.msu.edu), and EzTaxon (http://www.eztaxon.org) databases. The percent identity between the isolate from the patient and its closely related Sphingobacterium species of the 4 databases are shown in Table 1. The GenBank and RDP-II databases showed that the 16S rRNA gene sequence of the isolate from the patient was 100% homologous with that of S. spiritivorum strain NCTC 11386 (Accession number: GenBank, NR044077.1; RDP-II, S000752320). The EMBL and EzTaxon databases showed that the 16S rRNA gene sequence of the isolate from the patient was 100% homologous with that of S. spiritivorum strain ATCC 33861 (Accession number: EMBL, ACHA02000013; EzTaxon, ACHA01000008). The isolate from the patient showed percent identity of >99% with S. spiritivorum and >0.8% separation from other species. Thus, the isolate was confirmed to be S. spiritivorum [7].
For phylogenetic analysis, the resulting sequence was compared with those of reference strains of the most closely related Sphingobacterium species present in the GenBank databases. A phylogenetic tree was constructed by the neighbor-joining method using the Microseq 500 bp 16S rRNA sequences (Fig. 2).
Antimicrobial susceptibility was tested using the AST-N132 card from the Vitek 2 system (BioMérieux). The isolate was susceptible to cefepime, ciprofloxacin, levofloxacin, meropenem, minocycline, and trimethoprim-sulfamethoxazole; had moderate susceptibility to cefotaxime, ceftazidime, imipenem, and ticarcillin-clavulanic acid; but was resistant to amikacin, aztreonam, colistin, gentamicin, piperacillin, piperacillin-tazobactam, ticarcillin, and tobramycin.
The central venous catheter was removed. The antibiotic regimen was changed from cefepime to ciprofloxacin, because nephrotoxicity was suspected to be due to increased blood urea nitrogen and creatinine levels. After 3 days, the patient's fever subsided. Subsequent blood culture tests were negative for S. spiritivorum and any other microorganism. However, on the fifth day of ciprofloxacin therapy, she developed a fever again and her general condition worsened. On the eleventh day, she died of septic shock.
Sphingobacterium species are non-fermentative, non-motile, non-spore-forming aerobic gram-negative bacilli. They produce catalase, oxidase, and urease [1]. Sphingobacterium species grow on both BAP and MAC [2, 3]. Colonies are yellowish, circular, slightly convex, smooth, opaque, and non-hemolytic on BAP after 2 days of incubation [2, 3]. To date, 15 species, including S. anhuiense, S. antarcticus, S. bambusae, S. canadense, S. composti, S. daejeonense, S. faecium, S. heparinum, S. kitahiroshimense, S. multivorum, S. piscium, S. shayense, S. siyangense, S. spiritivorum, and S. thalpophilum, have been described in the genus Sphingobacterium [4, 8-17].
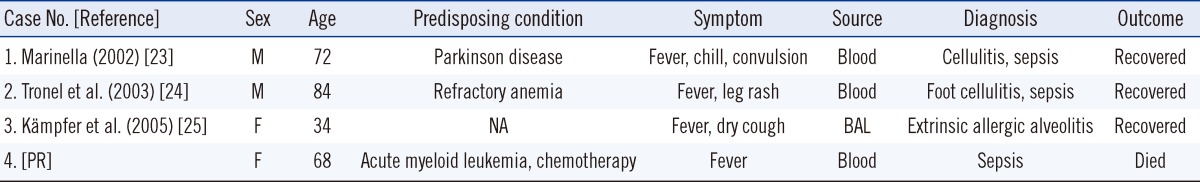
Sphingobacterium species are usually isolated from soil, water, and plant material, and only a few case reports of human infections caused by the species have been published [5]. Previously reported Sphingobacterium species isolated from human clinical specimens were S. multivorum and S. spiritivorum. To date, 7 cases of S. multivorum infection have been reported worldwide in relation to septicemia [5, 18-20], peritonitis [21], respiratory tract infection [22], and necrotizing fasciitis [23]. Only 3 cases of S. spiritivorum infection have been reported worldwide [24-26]. The present case and previously reported cases are compared in Table 2. In 2002, Marinella [24] first described a case of cellulitis-associated sepsis caused by an S. spiritivorum infection. In 2003, Tronel et al. [25] reported a case of S. spiritivorum bacteremia. In 2005, Kronel et al. [26] reported a case of cellulitis-associated sepsis caused by S. spiritivorum from the water reservoir of a steam iron.
In the present case, the patient was in an immunosuppressed condition due to the chemotherapy to treat acute myeloid leukemia. The patient was diagnosed with a catheter-related blood-stream infection because the time interval of positive blood culture signs from those between the peripheral vein and central venous catheter cultures was more than 2 hr. The source and transmission route of the S. spiritivorum infection in this case may have been a skin entry site of an intravascular device or a subcutaneous path of the catheter that had been in a close proximity with the natural habitats of this organism.
The 16S rRNA sequencing analysis can be a useful and definitive method particularly for the identification of clinically significant bacterial isolates with ambiguous biochemical profiles or of rarely encountered bacterial species [27, 28]. We confirmed the identity of the blood isolate, first identified biochemically as S. spiritivorum, by 16S rRNA sequencing analysis.
Sphingobacterium species are generally resistant to aminoglycosides and polymyxin B, but are susceptible to quinolones and trimethoprim-sulfamethoxazole in vitro. Susceptibility to β-lactam antibiotics is known to vary [1]. In 2009, Lambiase et al. [29] reported that 13 S. multivorum and 8 S. spiritivorum isolates from sputum samples in 332 patients with cystic fibrosis were resistant to aminoglycosides, but susceptible to quinolones and trimethoprim-sulfamethoxazole. They also found that S. multivorum isolates were resistant to all β-lactams, whereas the S. spiritivorum isolates were susceptible to ceftazidime, piperacillin, and carbapenems. The isolate of this case was susceptible to cefepime, meropenem, minocycline, as well as ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole.
This is the first reported case of human S. spiritivorum infection in Korea, which shows that S. spiritivorum can be a fatal human opportunistic pathogen in immunocompromised patients, despite human infection being rare.
References
1. Versalovic J, Carroll KC, editors. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press;2011. p. 723–727.
2. Holmes B, Owen RJ, Weaver RE. Flavobacterium multivorum, a new species isolated from human clinical specimens and previously known as group IIk, biotype 2. Int J Syst Bacteriol. 1981; 31:21–34.
3. Holmes B, Owen RJ, Hollis DG. Flavobacterium spiritivorum, a new species isolated from human clinical specimens. Int J Syst Bacteriol. 1982; 32:157–165.
4. Yabuuchi E, Kaneko T, Yano I, Moss CW, Miyoshi N. Sphingobacterium gen. nov., Sphingobacterium spiritivorum comb. nov., Sphingobacterium multivorum comb. nov., Sphingobacterium mizutae sp. nov., and Flavobacterium indologenes sp. nov.: Glucose-nonfermenting gram-negative rods in CDC groups IIK-2 and IIb. Int J Syst Bacteriol. 1983; 33:580–598.
5. Aydo an M, Yumuk Z, Dündar V, Arisoy ES. Sphingobacterium multivorum septicemia in an infant: Report of a case and review of the literature. Türk Mikrobiyol Cem Derg. 2006; 36:44–48.
6. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC;2008.
7. Clinical and Laboratory Standards Institute. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; Approved guideline. CLSI document MM18-A. Wayne, PA: Clinical and Laboratory Standards Institute;2008.
8. Wei W, Zhou Y, Wang X, Huang X, Lai R. Sphingobacterium anhuiense sp. nov., isolated from forest soil. Int J Syst Evol Microbiol. 2008; 58:2098–2101. PMID: 18768611.
9. Shivaji S, Ray MK, Rao NS, Saisree L, Jagannadham MV, Kumar GS, et al. Sphingobacterium antarcticus sp. nov., a psychrotrophic bacterium from the soils of Schirmacher oasis, Antarctica. Int J Syst Bacteriol. 1992; 42:102–106.
10. Duan S, Liu Z, Feng X, Zheng K, Cheng L. Sphingobacterium bambusae sp. nov., isolated from soil of bamboo plantation. J Microbiol. 2009; 47:693–698. PMID: 20127461.
11. Mehnaz S, Weselowski B, Lazarovits G. Sphingobacterium canadense sp. nov., an isolate from corn roots. Syst Appl Microbiol. 2007; 30:519–524. PMID: 17629434.
12. Yoo SH, Weon HY, Jang HB, Kim BY, Kwon SW, Go SJ, et al. Sphingobacterium composti sp. nov., isolated from cotton-waste composts. Int J Syst Evol Microbiol. 2007; 57:1590–1593. PMID: 17625199.
13. Kim KH, Ten LN, Liu QM, Im WT, Lee ST. Sphingobacterium daejeonense sp. nov., isolated from a compost sample. Int J Syst Evol Microbiol. 2006; 56:2031–2036. PMID: 16957095.
14. Takeuchi M, Yokota A. Proposals of Sphingobacterium faecium sp. nov., Sphingobacterium piscium sp. nov., Sphingobacterium heparinum comb. nov., Sphingobacterium thalpophilum comb. nov. and two geno-species of the genus Sphingobacterium and synonymy of Flavobacterium yabuuchiae and Sphingobacterium spiritivorum. J Gen Appl Microbiol. 1992; 38:465–482.
15. Matsuyama H, Katoh H, Ohkushi T, Satoh A, Kawahara K, Yumoto I. Sphingobacterium kitahiroshimense sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2008; 58:1576–1579. PMID: 18599698.
16. He X, Xiao T, Kuang H, Lan X, Tudahong M, Osman G, et al. Sphingobacterium shayense sp. nov., isolated from forest soil. Int J Syst Evol Microbiol. 2010; 60:2377–2381. PMID: 19933581.
17. Liu R, Liu H, Zhang CX, Yang SY, Liu XH, Zhang KY, et al. Sphingobacterium siyangense sp. nov., isolated from farm soil. Int J Syst Evol Microbiol. 2008; 58:1458–1462. PMID: 18523194.
18. Potvliege C, Dejaegher-Bauduin C, Hansen W, Dratwa M, Collart F, Tielemans C, et al. Flavobacterium multivorum septicemia in a hemodialyzed patient. J Clin Microbiol. 1984; 19:568–569. PMID: 6715525.
19. Freney J, Hansen W, Ploton C, Meugnier H, Madier S, Bornstein N, et al. Septicemia caused by Sphingobacterium multivorum. J Clin Microbiol. 1987; 25:1126–1128. PMID: 3597758.
20. Areekul S, Vongsthongsri U, Mookto T, Chettanadee S, Wilairatana P. Sphingobacterium multivorum septicemia: a case report. J Med Assoc Thai. 1996; 79:395–398. PMID: 8855615.
21. Dhawan VK, Rajashekaraiah KR, Metzger WI, Rice TW, Kallick CA. Spontaneous bacterial peritonitis due to a group IIk-2 strain. J Clin Microbiol. 1980; 11:492–495. PMID: 7381015.

22. Reina J, Borrell N, Figuerola J. Sphingobacterium multivorum isolated from a patient with cystic fibrosis. Eur J Clin Microbiol Infect Dis. 1992; 11:81–82. PMID: 1563393.
23. Grimaldi D, Doloy A, Fichet J, Bourgeois E, Zuber B, Wajsfisz A, et al. Necrotizing fasciitis and septic shock related to the uncommon gram-negative pathogen Sphingobacterium multivorum. J Clin Microbiol. 2012; 50:202–203. PMID: 22075581.
24. Marinella MA. Cellulitis and sepsis due to Sphingobacterium. JAMA. 2002; 288:1985. PMID: 12387649.
25. Tronel H, Plesiat P, Ageron E, Grimont PA. Bacteremia caused by a novel species of Sphingobacterium. Clin Microbiol Infect. 2003; 9:1242–1244. PMID: 14686992.
26. Kämpfer P, Engelhart S, Rolke M, Sennekamp J. Extrinsic allergic alveolitis (hypersensitivity pneumonitis) caused by Sphingobacterium spiritivorum from the water reservoir of a steam iron. J Clin Microbiol. 2005; 43:4908–4910. PMID: 16145174.
27. Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, et al. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol. 2003; 41:1996–2001. PMID: 12734240.

28. Arosio M, Nozza F, Rizzi M, Ruggeri M, Casella P, Beretta G, et al. Evaluation of the MicroSeq 500 16S rDNA-based gene sequencing for the diagnosis of culture-negative bacterial meningitis. New Microbiol. 2008; 31:343–349. PMID: 18843888.
29. Lambiase A, Rossano F, Del Pezzo M, Raia V, Sepe A, de Gregorio F, et al. Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res Notes. 2009; 2:262. PMID: 20030840.
Fig. 1
(A) Gram-negative bacilli from smear preparations of the positive blood cultures (Gram stain, ×1,000). (B) Yellow-colored colonies of Sphingobacterium spiritivorum on a blood agar plate.
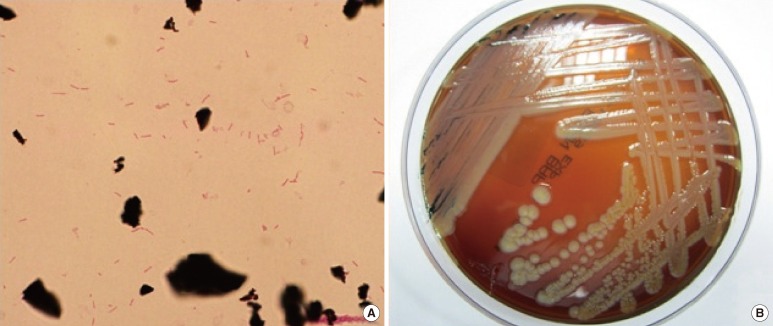
Fig. 2
Phylogenetic relationships of the isolate from the present patient and related Sphingobacterium species, constructed by the neighbor-joining method by using the Microseq 500 bp 16S rRNA sequences. All names and accession numbers are given as cited in the GenBank database. The tree was drawn with branch length as the evolutionary distances. The scale bar length of 0.01 indicates 1% sequence distance.
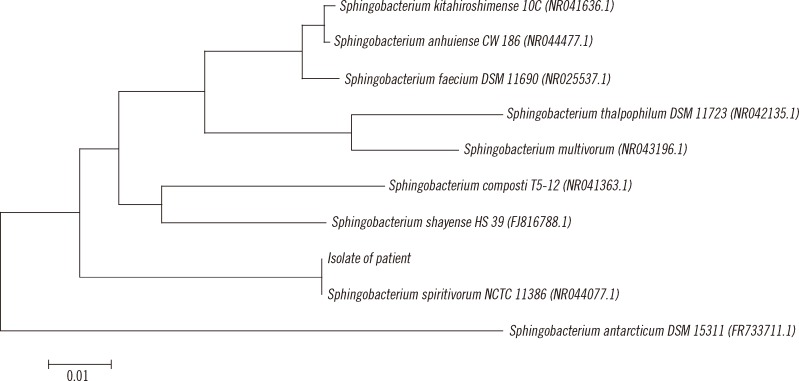
Table 1
Sequence comparison between the isolate from the patient and its most similar species
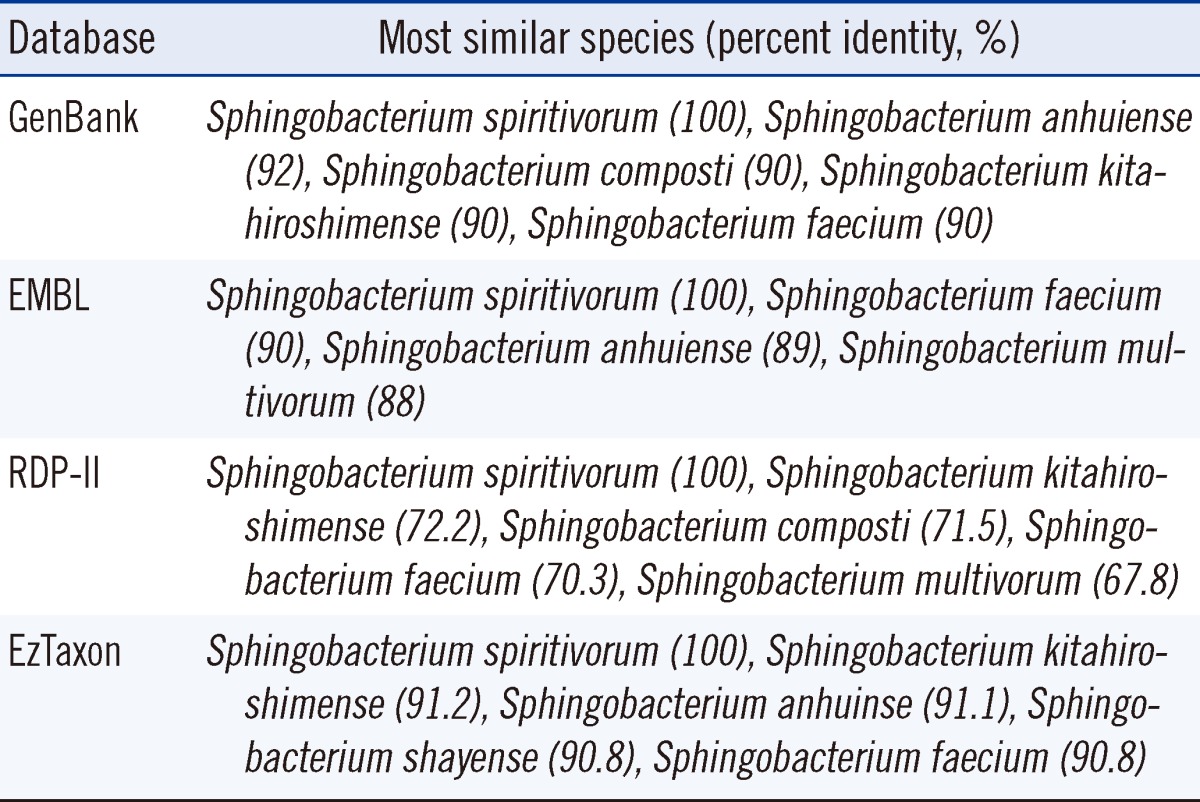
Database: GenBank (http://www.ncbi.nlm.nih.gov/genbank), EMBL (The European Molecular Biology Laboratory, http://www.ebi.ac.uk/embl), RDP-II (The Ribosomal Database Project, http://rdp.cme.msu.edu), and EzTaxon (http://www.eztaxon.org).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download