Abstract
Background
The primary purpose of this study was to investigate the prevalence and characteristics of p16 methylation and determine the prognostic implications of the clinical data, hematologic data, and p16 methylation changes in plasma cell myeloma (PCM).
Methods
We reviewed clinical characteristics and results of laboratory tests and investigated the response to combination chemotherapy and survival time. DNA methylation of the p16 gene was tested by methylation-specific PCR. Clinical significance was evaluated.
Results
A total of 103 patients were enrolled in this study. The median patient age was 59.0 yr at diagnosis and the male to female ratio was 1.15:1. According to the International Staging System (ISS), patients were diagnosed as stage: I (N=17, 16.5%), II (N=41, 39.8%), III (N=39, 37.9%), or not classified (N=6). Forty-five (43.7%) patients and 36 (35.0%) patients showed abnormal karyotype and complex karyotype, respectively, on the chromosome study. The p16 methylation was observed in 39 (37.9%) of 103 patients, but there was no significant association between p16 methylation status and other clinical or laboratory factors and survival outcome. Male gender, albumin, and complex karyotype were independent prognostic factors for overall survival according to multivariate analysis (P<0.05).
Plasma cell myeloma (PCM) is a clonal B-cell malignancy with a terminally differentiated plasma cell phenotype. The incidence rate is 5.6 per 100,000 persons per year and the median age at diagnosis is 70 yr [1, 2]. Recent improvements have been observed in patient clinical management, particularly with highdose therapy use followed by autologous stem cell transplantation. New drugs such as thalidomide, lenalidomide, bortezomib, and bisphosphonates, have also been developed. As a result, the 5-yr survival rate was 34% in 1996-2003, up from 25% in 1975-1977 [1, 3, 4]. The prognosis of PCM is highly variable, with survival ranging from a few days to more than 10 yr; however, PCM remains incurable. Therefore, it is essential to recognize clinical or biological parameters at diagnosis that can be used to predict patient outcome and to identify patients for whom aggressive therapy is indicated.
Previously identified prognostic factors include β2-microglobulin, serum albumin, hemoglobin, and cytogenetic aberrations [5-7], but the meaning and prognostic impact of methylation abnormalities are actively being studied. Aberrant methylation of CpG islands is one kind of epigenetic change observed in a wide range of cancers [8-10]. CpG islands of tumor suppressor genes are aberrantly methylated, resulting in transcriptional repression in many cancers, the effect of which is equivalent to mutation and deletions in carcinogenesis. The p16 tumor suppression gene is one of the most common genes, which is hypermethylated and detected in many cancers, including PCM [9, 11, 12]. The p16 genes encode cell cycle regulators involved in inhibiting G1 phase progression. Methylation of p16 genes has been linked with poor clinical outcome in bladder tumors, colorectal cancer, and lung cancer [13, 14]. However, the prognostic impact of p16 methylation in PCM is still unclear, and various results have been reported [8, 15].
The primary purpose of this study was to investigate the prevalence and characteristics of p16 methylation and to determine the prognostic implications of the clinical data, hematologic data, and p16 methylation changes in PCM.
Approval for this study was obtained from the Institutional Review Board of St. Mary's Hospital, The Catholic University of Korea (KC09EISI0393).
Between January 2004 and July 2009, 103 patients at St. Mary's Hospital, Seoul, Korea, with newly diagnosed PCM were enrolled. Diagnosis and staging were classified according to the WHO classification of Tumours of Haematopoietic and Lymphoid Tissue [2]. Clinical and laboratory characteristics of patients at diagnosis were collected from medical chart reviews. Analyzed characteristics included age, sex, percentage of plasma cells in bone marrow, hemoglobin level, white blood cell (WBC) and platelet counts, serum calcium, creatinine, lactate dehydrogenase (LDH), albumin, β2-microglobulin, immunoglobulin levels, serum/urine protein electrophoresis, serum and urine immunoelectrophoresis or immunofixation, and serum free light chain levels. Disease stages were classified according to the International Staging System (ISS) [16]. Responses to combination chemotherapy were defined according to International Myeloma Working Group uniform response criteria [17]. Immunofixation on the serum, urine, and bone marrow tests were conducted at follow-up to determine the treatment responses; survival times were determined by chart review.
Chromosome studies using a trypsin-Giemsa banding technique were performed on bone marrow cells at diagnosis. Metaphase cells were obtained from short-term unstimulated cultures, and at least 20 cells in metaphase were analyzed. A complex karyotype was defined as 3 or more chromosomal aberrations, including at least 1 structural aberration [18].
Methylation-specific PCR involves the chemical modification of genomic DNA using sodium bisulfate, which specifically converts cytosine to uracil in the unmethylated regions only. PCR using primers specific for both methylated DNA and modified DNA by sodium bisulfate can be used to determine the presence of methylated DNA in a given sample.
Bone marrow cells were scraped from bone marrow aspiration slides. DNA extractions were performed by QIAamp micro DNA kit, catalog number 56304 (QIAGEN GmbH, Hilden, Germany).
DNA concentrations were measured using a Nano-Drop 2000 (Thermo Fisher Scientific Inc., Wilmington, MA, USA) and adjusted to 500 ng/20 µL. Bisulfate treatment was performed using the EZ DNA Methylation Kit (Zymo Research Corporation, Orange, CA, USA). C/T conversion reagent was prepared by mixing 900 µL distilled water, 300 µL M-dilution buffer, and 50 µL dissolving buffer, and incubating for 10 min at room temperature. A 130-µL aliquot of C/T conversion reagent was added to 20 µL DNA and incubated for 10 min at 98℃, for 150 min at 64℃, and at 4℃ (hold). The 150-µL sample solution and 500 µL M-binding buffers were applied to an ion chromatography column, and then 200 µL M-desulphonation buffer was added to the column and incubated for 15-20 min at room temperature. M-elution buffer was added to the column, centrifuged at 10,000×g for 15 sec, and eluted. This bisulfate-treated DNA was used for PCR.
Methylation-specific PCR uses specific primers to assess methylation status for a given gene. Primers for p16 gene-promoter regions were designed according to a previous report [19]. Primer sequences were: methylated forward primer (p16-MF) 5'-TTATTAGAGGGTGGGGCGGATCGC-3', methylated reverse primer (p16-MR) 5'-GACCCCGAACCGCGACCGTAA-3', unmethylated forward primer (p16-UF) 5'-TTATTAGAGGGTGGGGTGGATTGT-3', unmethylated reverse primer (p16-UR) 5'-CAACCCCAAACCACAACCATAA-3'. Amplification was carried out in a C1000 thermal cycler (BIO-RAD; Hercules, CA, USA). PCR conditions were as follows: 1 cycle at 94℃ for 15 min; 35 cycles of 95℃ for 35 sec, 65℃ for 45 sec, and 72℃ for 40 sec; and 1 cycle of 72℃ for 5 min. DNA from the DLD-1 colon cancer cell line is reported to have a methylated p16 gene and negative expression of the gene by northern blot; hence, the DLD-1 cancer cell line was used as a positive control for monitoring DNA bisulfate modification and methylation-specific PCR [20]. Each PCR product was directly loaded onto a 2.5% agarose gel, stained with ethidium bromide, and directly visualized under UV illumination.
Independent sample t-test was used to assess the association among continuous variables, and the ANOVA was used between continuous variables and ISS stages. The chi-square test was applied between categorical variables and treatment responses. The independent sample t-test was used. Overall survival was the chosen end point (for any cause of death, disease, or other causes), and survivals were plotted on Kaplan-Meier curves and compared using the log-rank test. Prognostic factors for overall survival were determined using the Cox proportional hazard model for multivariate analysis. Statistical analyses were performed using MedCalc version 11.2 (MedCalc Software, Mariakerke, Belgium). Statistical significance was assumed at a two-sided P value of <0.05.
The patients included 55 men and 48 women (the male to female ratio was 1.15:1, and the median±SD [range] age was 59.0±9.9 [34-80] yr at diagnosis). Patient disease staging perform ed according to the ISS was as follows: stage I (N=17, 16.5%); stage II (N=41, 39.8%); and stage III (N=39, 37.9%). Six patients were not classified because β2-microglobulin data was missed at diagnosis. The types of M protein identified were IgG, 49 (47.6%); IgA, 30 (29.1%); IgD, 3 (2.9%); and light chain disease, 21 (20.4%). Forty-five (43.7%) and 36 (35.0%) patients showed an abnormal karyotype and complex karyotype on chromosome study, respectively. The responses to combination chemotherapy were next complete response (CR), 41 (45.1%); very good partial response (VGPR), 15 (16.5%); partial response (PR), 20 (22.0%); and stable disease (SD), 15 (16.5%). The responses of 12 patients could not be determined due to follow-up loss (Table 1).
Thirty-nine (37.9%) of the 103 patients showed p16 methylation. Positive rates of p16 methylation were 35.3%, 36.6%, and 41.0% for ISS I, II, and III, respectively (P=0.887). p16 methylation was detected in 34.5% (20/58) and 42.2% (19/45) of the patients with normal karyotypes and abnormal karyotypes, respectively (P=0.55). p16 methylation was not significantly associated with major cytogenetic abnormalities in PCM including complex karyotype, 14q32 abnormalities, t(11;14)(q13,q32), t(4;14)(p16;q32), and del(17p13) (P>0.05). Additionally, other laboratory and clinical factors were not significantly associated with p16 methylation status.
The ISS and Durie & Salmon stage and other factors were assessed by univariate analysis for survival to identify significant prognostic effects. Based on the univariate analysis, gender, creatinine, platelet, hemoglobin, albumin β2-microglobulin, complex karyotype, ISS, and the Durie & Salmon stage were significant prognostic factors for overall survival (P<0.05). The male gender, low serum albumin level, and complex karyotype were significant independent poor prognostic factors for overall survival based on the final multivariate model obtained by stepwise selection of variables (P<0.05) (Table 2).
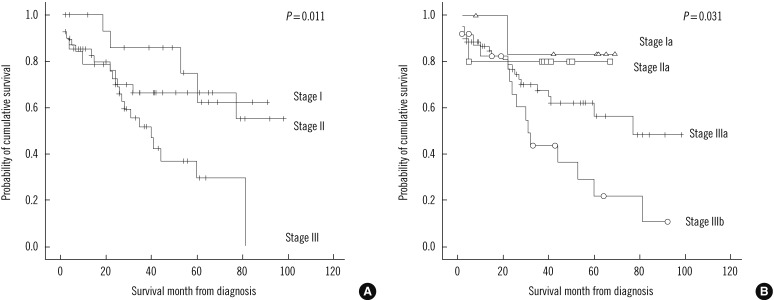
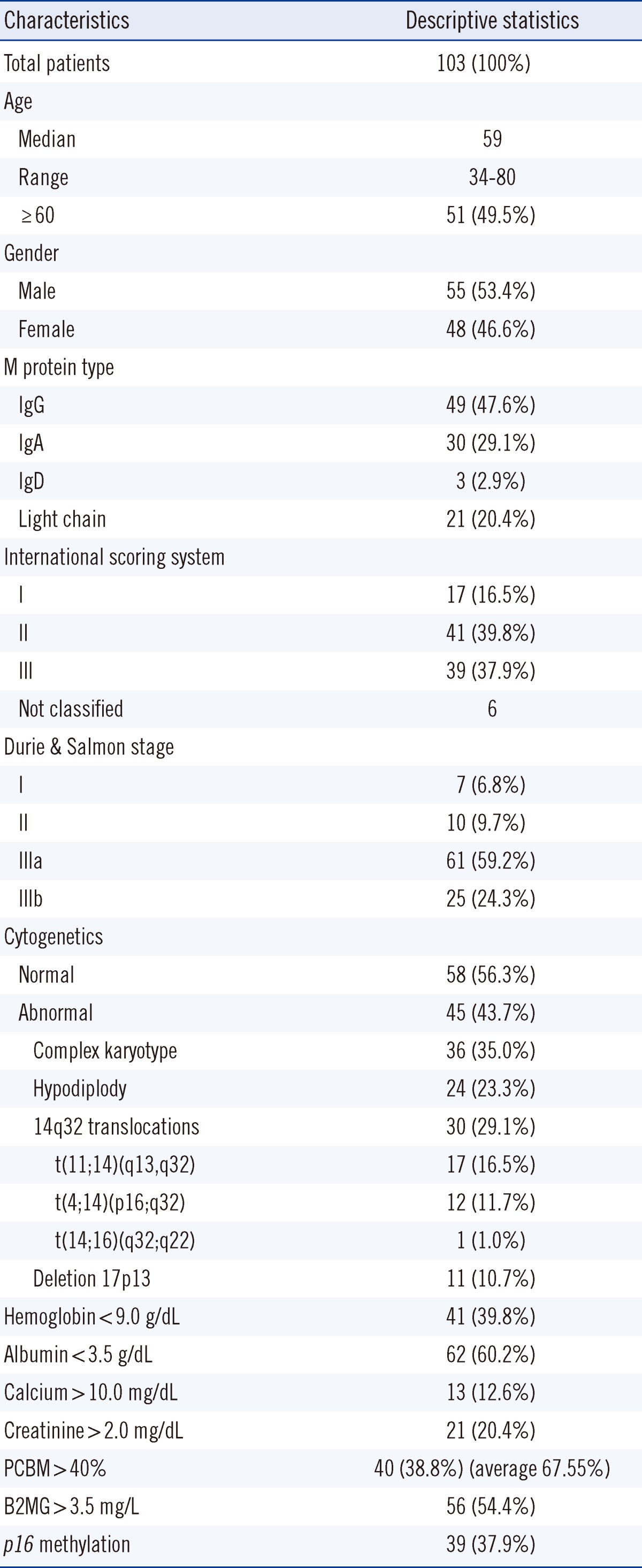
The median follow-up duration was 31.5 (1-98) months (minimum-maximum, 0-98 months). Thirty-nine (47.6%) of the 82 patients died during the study. Patients in stage III of the ISS categories showed significantly different overall survival (P=0.012). Patients in the stage IIIb of Durie & Salmon stage also showed significant stratifications of patients according to the overall survival (P=0.012) (Fig. 1 and Table 2).
This study analyzed clinical and laboratory data and p16 methylation in 103 cases with PCM in a single Korean institution. The median age in the present study was 59 yr, slightly less than the previously reported median age (70 yr) of PCM, with a minor male predominance. M protein types showed a relatively higher incidence of IgA type than that reported previously (20%), and the incidence of other M protein types were similar to those in previous studies [2].
Cytogenetic status has become an important prognostic indicator for patients with PCM. A single genetic factor such as hyperdiploidy, t(11;14) or hypodiploidy, t(4;14), t(14;16) are known prognostic factors [6]. A complex karyotype was shown to be an independent prognostic factor in this study. This result indicates that overall genetic instability should be considered in estimating PCM with previously known single genetic factors.
Aberrant gene promoter methylation is a common phenomenon, although the meaning and prognostic impact on survival of methylation in PCM has not been characterized. The frequency of p16 methylation was shown to be 10-60% of PCM patients in recent studies [15, 21-23]. In this study, 39 (37.9%) of the 103 patients were positive for p16 methylation, and positive rates of p16 methylation increased according to ISS stage. These differences may have been caused by the differences in type and num ber of primers used, detection methods, and the number of plasma cells in the sample. Previous studies using 2 primer sets revealed a higher p16 methylation rate than that revealed by our results [15, 21]. Quantitative analysis using purified plasma cells is necessary to accurately detect p16 methylation in PCM.
Normal subjects were shown to have no p16 methylation in a previous study, which investigated p16 methylation by bisulfate direct sequencing in 20 healthy donor samples [24]. In the present study, p16 methylation was detected in 34.5% of the normal karyotype group. These findings imply that epigenetic changes, including p16 methylation, play a role in tumorigenesis of PCM. However, there is no significant difference in p16 methylation between ISS stages, abnormal karyotype, and complex karyotype in this study. Additionally, overall survival did not differ significantly between patients with and without p16 methylation. However, previous studies reported that p16 methylation is associated with survival in Korean PCM patients [15]. Similarly to this study, it was reported that p16 methylation does not influence survival outcome [8, 22]. To demonstrate the significance and prognostic impact of epigenetic changes on PCM, methylation of various genes, quantitative analysis, and their overall effect should be examined.
Male gender, hemoglobin, platelet, creatinine, albumin, β2-microglobulin, and complex karyotype are significantly associated with survival time according to univariate analysis. According to multivariate analysis, male gender, albumin, and complex karyotype were independent factors for survival time. Poor outcomes of the male patients in the present study differs from previously known prognostic factors [6]. The most commonly used classification was developed by Durie and Salmon in 1975 [25]. Despite the general use of the Durie and Salmon staging system, there is no universal agreement on its prognostic value. The ISS is reproducible in all age groups, different geographic origins, and treatments; hence, it can be used to predict survival [16, 25-27]. Durie and Salmon staging and ISS showed a similar prognostic impact in this study (Fig. 1 and Table 2). The overall survival of the highest stage (III or IIIb) of both systems were significantly different for stage I PCM in univariate analysis (stage III [P=0.012] in ISS, stage IIIb [P=0.012] in Durie and Salmon system).
We analyzed the prognostic impact of each clinical factor, laboratory factors, and p16 methylation status on overall survival. The male gender, low serum albumin level, and complex karyotype were significant independent poor prognostic factors for PCM. A single p16 methylation status may play a role in tumorigenesis of PCM, but did not have a prognostic impact.
Acknowledgement
This study was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Korea (SN: A092258).
References
1. Ries LAG MD KM, Stinchcomb DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, Lewis DR, Clegg L, Eisner MP, Reichman M, Edwards BK, editors. SEER Cancer Statistics Review, 1975-2005. National Cancer Institute;http://seer.cancer.gov/csr/1975_2005/(based on November 2007 SEER data submission, posted to the SEER web site 2008).
2. McKenna RW, Kyle RA, Kuehl WM, Grogan TM, Harris NL, Coupland RW. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. Plasma cell neoplasm. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 2008. 4th ed. Lyon: IARC;p. 200–213.
3. Brenner H, Gondos A, Pulte D. Expected long-term survival of patients diagnosed with multiple myeloma in 2006-2010. Haematologica. 2009; 94:270–275. PMID: 19144659.

4. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008; 58:71–96. PMID: 18287387.

5. Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood. 2007; 109:3489–3495. PMID: 17209057.
6. Bladé J, Rosiñol L, Cibeira MT. Prognostic factors for multiple myeloma in the era of novel agents. Ann Oncol. 2008; 19(Suppl 7):vii117–vii120. PMID: 18790932.

7. Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C. Group Français de Cytogénétique Hématologique. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001; 98:2229–2238. PMID: 11568011.

8. Gonzalez-Paz N, Chng WJ, McClure RF, Blood E, Oken MM, Van Ness B, et al. Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications. Blood. 2007; 109:1228–1232. PMID: 16840723.
9. Goto T, Mizukami H, Shirahata A, Sakata M, Saito M, Ishibashi K, et al. Aberrant methylation of the p16 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009; 29:275–277. PMID: 19331161.
10. Ohashi H, Tsushita K, Utsumi M, Shimoyama M, Murate T, Uchida T, et al. Relationship between methylation of the p15 gene and ectopic expression of the EVI-1 gene in myelodysplastic syndromes (MDS). Leukemia. 2001; 15:990–991. PMID: 11417490.
11. Guillerm G, Gyan E, Wolowiec D, Facon T, Avet-Loiseau H, Kuliczkowski K, et al. p16INK4a and p15INK4b gene methylations in plasma cells from monoclonal gammopathy of undetermined significance. Blood. 2001; 98:244–246. PMID: 11418489.
12. Guzman LM, Koriyama C, Akiba S, Eizuru Y, Castillo D, Corvalan A, et al. High frequency of p16 promoter methylation in non-small cell lung carcinomas from Chile. Biol Res. 2007; 40:365–372. PMID: 18449464.

13. Dominguez G, Silva J, Garcia JM, Silva JM, Rodriguez R, Muñoz C, et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003; 530:9–17. PMID: 14563526.
14. Tanaka R, Wang D, Morishita Y, Inadome Y, Minami Y, Iijima T, et al. Loss of function of p16 gene and prognosis of pulmonary adenocarcinoma. Cancer. 2005; 103:608–615. PMID: 15612080.
15. Park G, Kang SH, Lee JH, Suh C, Kim M, Park SM, et al. Concurrent p16 methylation pattern as an adverse prognostic factor in multiple myeloma: a methylation-specific polymerase chain reaction study using two different primer sets. Ann Hematol. 2011; 90:73–79. PMID: 20721556.
16. Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Bladé J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005; 23:3412–3420. PMID: 15809451.

17. Durie BG, Harousseau JL, Miguel JS, Bladé J, Barlogie B, Anderson K, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006; 20:1467–1473. PMID: 16855634.

18. Göhring G, Michalova K, Beverloo HB, Betts D, Harbott J, Haas OA, et al. Complex karyotype newly defined: the strongest prognostic factor in advanced childhood myelodysplastic syndrome. Blood. 2010; 116:3766–3769. PMID: 20802024.

19. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996; 93:9821–9826. PMID: 8790415.

20. Zheng S, Chen P, McMillan A, Lafuente A, Lafuente MJ, Ballesta A, et al. Correlations of partial and extensive methylation at the p14(ARF) locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis. 2000; 21:2057–2064. PMID: 11062168.

21. Martin P, Garcia-Cosio M, Santon A, Bellas C. Aberrant gene promoter methylation in plasma cell dyscrasias. Exp Mol Pathol. 2008; 84:256–261. PMID: 18410922.

22. Ribas C, Colleoni GW, Felix RS, Regis Silva MR, Caballero OL, Brait M, et al. p16 gene methylation lacks correlation with angiogenesis and prognosis in multiple myeloma. Cancer Lett. 2005; 222:247–254. PMID: 15863274.
23. Yuregir OO, Yurtcu E, Kizilkilic E, Kocer NE, Ozdogu H, Sahin FI. Detecting methylation patterns of p16, MGMT, DAPK and E-cadherin genes in multiple myeloma patients. Int J Lab Hematol. 2010; 32:142–149. PMID: 19302404.
24. Shiraz OB, Galehdari H, Yavarian M, Geramizadeh B. Possible down regulation of the p16 gene promoter in individuals with hepatocellular carcinoma. Hepat Mon. 2011; 11:719–723. PMID: 22235214.

25. Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975; 36:842–854. PMID: 1182674.

26. Hari PN, Zhang MJ, Roy V, Pérez WS, Bashey A, To LB, et al. Is the International Staging System superior to the Durie-Salmon staging system? A comparison in multiple myeloma patients undergoing autologous transplant. Leukemia. 2009; 23:1528–1534. PMID: 19322205.

27. Harousseau JL, Dreyling M. Multiple myeloma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2008; 19(Suppl 2):ii55–ii57. PMID: 18456769.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download