Abstract
Acinetobacter baumannii is an important microorganism responsible for a number of nosocomial outbreaks, in particular, in intensive care units (ICUs). We investigated a nosocomial infection caused by multidrug-resistant (MDR) A. baumannii in a neonatal intensive care unit (NICU) in Korea. A. baumannii isolates were characterized using Etest (AB Biodisk, Sweden), two multiplex PCR assays, and multilocus sequence typing (MLST) scheme. PCR and PCR mapping experiments were performed for detecting and characterizing the determinants of antimicrobial resistance. Eight strains isolated from an NICU belonged to European (EU) clone II and revealed only one sequence type (ST), namely, ST357. All the isolates were susceptible to imipenem but were resistant to amikacin, gentamicin, ceftazidime, cefepime, and ciprofloxacin. To the best of our knowledge, this is the first report of a nosocomial infection in an NICU in Korea caused by ST357 MDR/carbapenem-susceptible A. baumannii strains. This result demonstrates that nosocomial outbreaks of MDR/carbapenem-susceptible strains as well as MDR/carbapenem-resistant isolates may occur in NICUs.
Acinetobacter baumannii, a ubiquitous aerobic, glucose-non-fermenting, gram-negative bacterium, is able to survive on inanimate objects or fomites and colonize the human body. During the last 2 decades, this microorganism has been the etiological agent of serious opportunistic infections and nosocomial outbreaks [1]. In particular, nosocomial infections and colonization of multidrug-resistant (MDR) A. baumannii strains have been reported globally, mainly in intensive care units (ICUs). Most MDR A. baumannii strains are highly resistant to many antimicrobial agents, and therefore, therapeutic options are increasingly limited. Because of these reasons, it should be emphasized that preventing and controlling the dissemination of these strains is very important [2-4]. A. baumannii is not only a nosocomial pathogen in adults, but it is also responsible for neonatal infections, generally causing pneumonia [5].
Although numerous nosocomial infections caused by MDR A. baumannii strains are appearing rapidly worldwide, especially in the ICUs, there is a relative paucity of data associated with its dissemination in neonatal ICUs (NICUs) in Korea. In the present study, we aimed to determine the clonal relationships among clinical isolates obtained from an NICU of a university hospital in Daejeon, Korea. In addition, the genetic basis for multidrug resistance was investigated with special focus on carbapenemases, aminoglycoside-modifying enzymes (AMEs), 16S rRNA methylase, and mutations in the gyrA and parC genes.
A total of 8 consecutive and non-duplicate MDR A. baumannii strains were collected from the NICU of the university hospital in Daejeon, Korea, during the period from October 2011 to February 2012 (Table 1). A. baumannii strains were identified using the Vitek 2 automated instrument ID system (BioMérieux, Marcy l'Etoile, France) and by sequencing the partial rpoB housekeeping gene, as described previously [6].
The minimum inhibitory concentrations (MICs) for A. baumannii isolates of amikacin, gentamicin, ceftazidime, cefepime, imipenem, and ciprofloxacin were determined using Etest (AB Biodisk, Solna, Sweden). The results obtained were interpreted as per the criteria approved by the CLSI guidelines [7]. Escherichia coli ATCC 25922 was used as a reference strain. Whole cell (genomic) DNA for PCR templates was obtained from each target strain by using a genomic DNA purification kit (SolGent, Daejeon, Korea), according to the manufacturer's instructions. Two multiplex PCR assays were used as previously described [8] to identify members of European (EU) clones I and II. The Oxford multilocus sequence typing (MLST) scheme [9], which uses 7 housekeeping genes (gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD24), was used to determine the sequence types (STs). An ST number was assigned by comparing the allele sequences to those in the MLST database (http://pubmlst.org/abaumannii/). Epidemiological typing of isolates was performed by repetitive extragenic palindromic sequence (REP)-PCR [10].
All A. baumannii isolates were subjected to PCR and sequencing assays for detecting the determinants of antimicrobial resistance and for identifying the mutations associated with fluoroquinolone resistance. Specific primers and PCR conditions for detection of antimicrobial resistance determinants were as described in a previous study [11]. The amplicons were purified with a PCR purification kit (SolGent) and sequenced using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3730XL DNA analyzer (PE Applied Biosystems).
A total of 8 A. baumannii isolates were identified on the basis of their rpoB gene analysis. Although all the isolates were susceptible to imipenem, they showed resistance to amikacin, gentamicin, ceftazidime, cefepime, and ciprofloxacin (Table 2). These isolates belonged to EU clone II and carried allele 66 of the intrinsic blaOXA-51-like genes, which agrees with their assignment to EU clone II. They were all identified as ST357 (1-12-3-2-2-145-3) by MLST analysis. To determine the clonality of all 8 A. baumannii isolates, REP-PCR was carried out using genomic DNA. They all displayed the same REP-PCR type (Fig. 1). Our results suggest that ST357 A. baumannii strains isolated during instances of nosocomial infections or colonization in the NICU of the university hospital in Daejeon, Korea, belonged to EU clone II. Nosocomial infections or colonization due to ST357 A. baumannii isolates have not been detected in Korea previously. Thus, to the best of our knowledge, this is the first report of ST357 MDR/carbapenem-susceptible A. baumannii strains causing NICU nosocomial infections in Korea. This result suggests that not only MDR/carbapenem-resistant strains but also MDR/carbapenem-susceptible isolates act as major causes of nosocomial outbreaks, especially in the NICU, in Korea.
To define the genetic basis of MDR A. baumannii isolates by detecting antimicrobial resistance determinants, PCR and PCR mapping experiments were performed. All 8 A. baumannii isolates had the combination of AMEs encoded by aac(6')-Ib/aph(3')-Ia and 16S rRNA methylase encoded by armA. They showed high-level resistance to amikacin (MIC ≥256 mg/L) and gentamicin (MIC ≥1,024 mg/L). Unlike AMEs, which varies in their substrate ranges, 16S rRNA methylases confer high-level resistance to almost all clinically important aminoglycosides [12, 13]. In addition, they all had sense mutations at the 83rd residue (serine to leucine) in gyrA and at the 80th residue (serine to leucine or tryptophan) in parC and had high-level ciprofloxacin resistance (MIC50 ≥32 mg/L). A major mechanism of fluoroquinolone resistance in gram-negative bacteria involves changes in the structure of DNA gyrase and DNA topoisomerase IV [14]. Notably, amino acid substitutions in both the GyrA and ParC polypeptides are consistent with a high-level fluoroquinolone-resistant phenotype [15]. In this study, all 8 isolates harbored sense mutations in both gyrA and parC and showed high-level resistance to ciprofloxacin. In contrast, carbapenem-resistance genes, including metallo-β-lactamase genes and blaOXA-23-, blaOXA-24-, and blaOXA-58-like genes, were not found in this study.
Carbapenems are frequently the drug of choice to treat A. baumannii infections, and carbapenem resistance is, in itself, sufficient to define a highly resistant phenotype. Although nosocomial infections and colonization due to MDR A. baumannii have been reported worldwide, previous studies were focused primarily on carbapenem-resistant strains [12, 16]. Consequently, MDR/carbapenem-susceptible isolates have been rarely recovered worldwide. In our study, ST357 MDR/carbapenem-susceptible strains were isolated from the NICU. Our results emphasize that investigations should be performed not only on carbapenem-resistant but also on carbapenem-susceptible MDR A. baumannii isolates for studying the nosocomial infections and outbreaks caused by this organism.
References
1. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008; 21:538–582. PMID: 18625687.
2. Wisplinghoff H, Edmond MB, Pfaller MA, Jones RN, Wenzel RP, Seifert H. Nosocomial bloodstream infections caused by Acinetobacter species in United States hospitals: clinical features, molecular epidemiology, and antimicrobial susceptibility. Clin Infect Dis. 2000; 31:690–697. PMID: 11017817.
3. Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977-2000. Infect Control Hosp Epidemiol. 2003; 24:284–295. PMID: 12725359.
4. Garnacho-Montero J, Ortiz-Leyba C, Fernández-Hinojosa E, Aldabó-Pallás E, Cayuela A, Marguez-Vácaro E, et al. Acinetobacter baumannii ventilator-associated pneumonia: epidemiological and clinical findings. Intensive Care Med. 2005; 31:649–655. PMID: 15785929.
5. Touati A, Achour W, Cherif A, Hmida HB, Afif FB, Jabnoun S, et al. Outbreak of Acinetobacter baumannii in a neonatal intensive care unit: antimicrobial susceptibility and genotyping analysis. Ann Epidemiol. 2009; 19:372–378. PMID: 19364663.
6. Gundi VA, Dijkshoom L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009; 155:2333–2341. PMID: 19389786.
7. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Fifteenth Informational supplement, M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute;2010.
8. Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007; 13:807–815. PMID: 17610600.
9. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005; 43:4382–4390. PMID: 16145081.
10. Bou G, Cerveró G, Domínguez MA, Quereda C, Martínez-Beltrán J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000; 6:635–643. PMID: 11284921.
11. Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrug-resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010; 30:498–506. PMID: 20890082.
12. Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–190. PMID: 19361944.
13. Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, et al. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010; 29:1349–1353. PMID: 20614151.

14. Akasaka T, Tanaka M, Yamaguchi A, Sato K. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob Agents Chemother. 2001; 45:2263–2268. PMID: 11451683.
15. Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008; 46:2499–2507. PMID: 18524965.
16. McGrath EJ, Chopra T, Abdel-Haq N, Preney K, Koo W, Asmar BI, et al. An outbreak of carbapenem-resistant Acinetobacter baumannii infection in a neonatal intensive care unit: investigation and control. Infect Control Hosp Epidemiol. 2011; 32:34–41. PMID: 21091204.
Fig. 1
Repetitive element sequence-based (REP)-PCR patterns of multidrug-resistant A. baumannii strains. Lane M, 1-kb DNA size marker; Lanes 1, 2, 3, 4, 5, 6, 7, and 8, isolates C10, C51, C64, C81, C84, C85, C132, and C147, respectively.
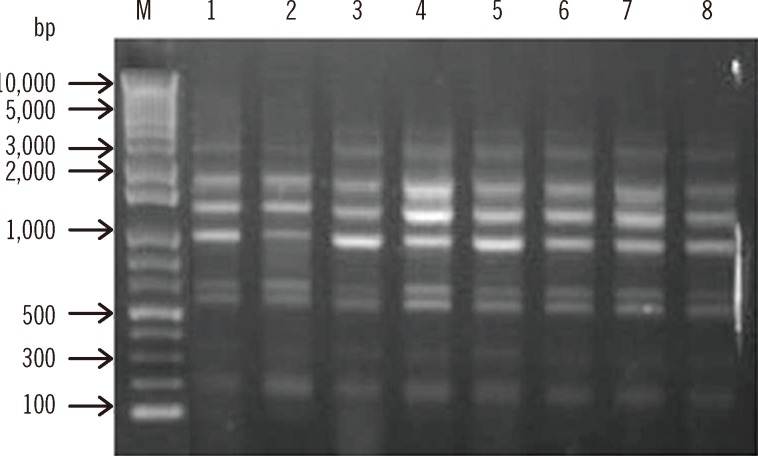
Table 1
Clinical characteristics of 8 patients infected with multidrug-resistant A. baumannii strains isolated in a neonatal intensive care unit
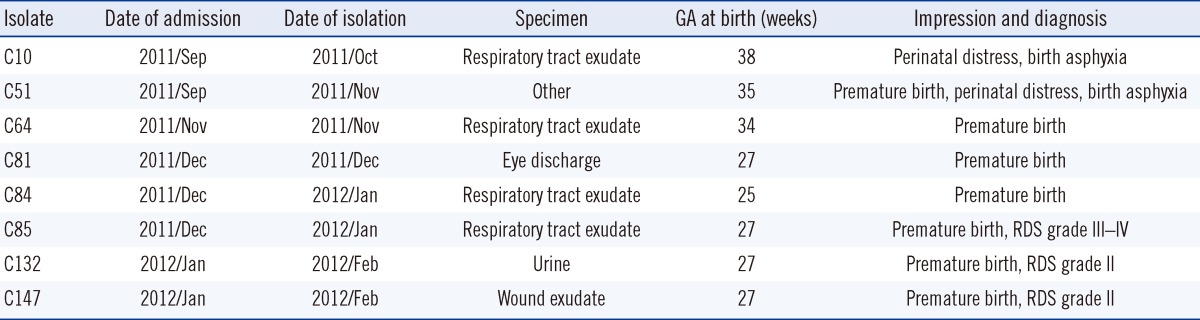
Table 2
Properties of multidrug-resistant A. baumannii strains isolated from a neonatal intensive care unit

*Indicates sense mutations at the 83rd residue (serine to leucine) in gyrA and at the 80th residue (serine to leucine or tryptophan) in parC.
Abbreviations: ST, sequence type; AMK, amikacin; GEN, gentamicin; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; CIP, ciprofloxacin; AMEs, aminoglycoside-modifying enzymes.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download