Abstract
Background
We evaluated the performance of three chromogenic media (Brilliance agar I [Oxoid, UK], Brilliance agar II [Oxoid], and ChromID MRSA [Biomérieux, France]) combined with broth enrichment and the Xpert MRSA assay for screening of methicillin-resistant Staphylococcus aureus (MRSA).
Methods
We obtained 401 pairs of duplicate nasal swabs from 321 patients. One swab was suspended overnight in tryptic soy broth; 50-µL aliquots of suspension were inoculated on the three chromogenic media. Brilliance agar I and II were examined after 24 hr, and ChromID MRSA, after 24 and 48 hr. The paired swab was processed directly using real-time PCR-based Xpert MRSA assay.
Results
True positives, designated as MRSA growth in any of the culture media, were detected with the prevalence of 17% in our institution. We report the sensitivity, specificity, positive predictive value, and negative predictive value of MRSA growth as follows: 92.3%, 94.0%, 75.9%, and 98.4% in Brilliance agar I (24 hr); 92.7%, 97.9%, 90.0%, and 98.5% in Brilliance agar II (24 hr); 95.6%, 95.8%, 82.3%, and 99.1% in ChromID MRSA (24 hr); 100%, 92.5%, 73.1%, and 100% in ChromID MRSA (48 hr); 92.6%, 96.7%, 85.1%, and 98.5% in Xpert MRSA assay. The agreement between the enriched culture and Xpert MRSA assay was 96.0%.
Conclusions
Three chromogenic culture media combined with enrichment and Xpert MRSA assay demonstrated similar capabilities in MRSA detection. The Xpert MRSA assay yielded results comparable to those of culture methods, saving 48-72 hr, thus facilitating earlier detection of MRSA in healthcare settings.
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the important nosocomial pathogens worldwide [1]. Patients carrying MRSA are at a greater risk for developing an S. aureus infection, compared with methicillin-susceptible S. aureus (MSSA)-colonized or non-colonized patients in the intensive care unit (ICU) [2]. Active surveillance and isolation policy have been recommended for controlling the spread of MRSA in healthcare facilities [3]. In Korea, S. aureus is the second most common isolate (16.6%) among hospital isolates, and the methicillin-resistance rate of S. aureus was about 69%, according to the 2009 KONSAR study [4]. A timely, affordable, and reliable screening method for MRSA is essential to improve hospital infection control, and there is a continued debate about the turn-around time, cost per test, and cost benefit.
The Xpert MRSA assay (Cepheid, Sunnyvale, CA, USA) is a real-time PCR assay that targets the staphylococcal cassette chromosome mec (SCCmec)-orfX junction created by incorporation of the genetic element carrying mecA into the S. aureus chromosome [5, 6]. The assaycan detect, within only 2 hr, strains with all SCCmec types, including SCCmec I, II, III, IVa, V, and VI found in healthcare-acquired (HA) and community-acquired (CA) MRSA [7, 8]. Currently, various selective and differential chromogenic media are available that allow direct colony color-based identification of the pathogen from the primary culture 18-24 hr after incubation. However, broth enrichment and incubation for 48 hr are necessary to increase the detection sensitivity, especially in cases with low bacterial density [9, 10].
In this study, we compared the clinical performance of three chromogenic media (Brilliance agar I [Oxoid Ltd., Cambridge, UK], Brilliance agar II [Oxoid], and ChromID MRSA [Biomérieux, Lyon, France]) combined with broth enrichment and the Xpert MRSA assay for MRSA detection by using nasal swabs from ICU patients.
This study was performed during March-May 2010. A total of 401 nasal samples were collected consecutively from 321 patients admitted to the ICU. The screening for MRSA was carried out on the first day in the ICU (321 initial samples) and was repeated weekly in 55 patients (80 follow-up samples) with prolonged hospitalization. The paired swabs (Copan, Brescia, Italy) were collected from each nostril of the same patient, respectively. The swab pair was transported at room temperature, and each swab was randomly used in the following process within 2 hr of collection.
One of the paired swabs was suspended in tryptic soy broth (TSB) (Becton, Dickinson and Company, Sparks, MD, USA) with 6.5% NaCl; after overnight incubation, 50 µL aliquots of suspension were inoculated on Brilliance agar I, Brilliance agar II, and ChromID MRSA. The two Brilliance agars were incubated for 24 hr (Brilliance agar I [24 hr] and Brilliance agar II [24 hr], respectively), and they were examined according to the manufacturer's recommendation. ChromID MRSA was incubated for 48 hr, and it was then examined at 24 and 48 hr (ChromID MRSA [24 hr] and ChromID MRSA [48 hr], respectively) by two investigators. Potential colonies were captured and identified as S. aureus if they tested positive for catalase production and latex agglutination tests (Eiken Chemical Co., Tokyo, Japan). Methicillin resistance was confirmed by oxacillin (1 µg) (Becton, Dickinson and Company) and cefoxitin (30 µg) (Becton, Dicksinson, and Company) disk diffusion susceptibility tests, according to the Clinical and Laboratory Standards Institute guideline [11]. The remaining TSB was frozen at -70℃ for further evaluation. A true-positive culture was defined as growth with phenotypic features compatible with MRSA and was affirmed by confirmatory testing (catalase and latex agglutination tests). A false-positive culture was defined as growth with phenotypic features compatible with MRSA, but was affirmed as non-MRSA by confirmatory testing.
The second swab of the pair was processed directly using the GeneXpert system (Cepheid) with Xpert MRSA cartridge (Cepheid) according to the manufacturer's instruction. In brief, the swab was placed in the elution reagent (black cap), vortexed at high speed for 10 sec, and dispensed into the port "S" of the Xpert MRSA cartridge. The cartridge was inserted and subjected to real-time PCR in the GeneXpert system, and the result was reported by the GeneXpert software (Cepheid). If the MRSA threshold (CT) value of the sample was ≤36, it was positively identified as MRSA.
A true-positive result, which was used as a reference result, was presumed as growth of MRSA in any one of the enriched chromogenic media. The sensitivity (%), specificity (%), positive predictive value (PPV, %), and negative predictive value (NPV, %) of each method was determined. For cases showing discrepant results between Xpert MRSA assay and cultures, both tests were repeated using the remaining TSB.
The CT values between two groups (initial screening samples versus follow-up screening samples; concordant samples versus discordant samples) were compared using the one-way ANOVA test (SPSS 13.0; SPSS Inc., Chicago, IL, USA). The threshold for statistical significance was a P value <0.05.
MRSA was identified in 68 (68/401, 17.0%) samples in at least one of the culture media. The prevalence of MRSA was 13.4% (43/321) for initial screening and 31.3% (25/80) for follow-up screening samples. The comparison of four screening methods is summarized in Table 1. The sensitivity was highest with ChromID MRSA (48 hr) (68/68, 100%), followed by ChromID MRSA (24 hr) (65/68, 95.6%), and lowest in both Brilliance agar I (24 hr) and Brilliance agar II (24 hr) (63/68, 92.6%). The specificity and PPV was highest with Brilliance agar II (24 hr) (95.8% and 90.0%, respectively) of all the culture media. NPVs were ≥98% in all the culture methods.
There were 25, 14, 20, and 7 samples in which growth on ChromID MRSA (48 hr), ChromID MRSA (24 hr), Brilliance agar I (24 hr), and Brilliance agar II (24 hr), respectively, was initially mistaken for MRSA, resulting in false-positive findings. The implicated isolates were mainly Enterococcus spp. (15/25), followed by coagulase-negative Staphylococcus (4/25) in ChromID MRSA; Bacillus spp. (8/20), followed by coagulase-negative Staphylococcus (5/20) and methicillin-susceptible S. aureus (4/20) in Brilliance agar I; and methicillin-resistant coagulase-negative Staphylococcus (4/7) and methicillin-susceptible S. aureus (3/7) in Brilliance agar II.
The Xpert MRSA assay showed positive results for 74 samples. Moreover, when growth of MRSA on any of the enriched culture media was designated as a reference method, the total agreement with the culture was 96.0% (385/401) in the initial analysis. The sensitivity, specificity, PPV, and NPV of the Xpert MRSA assay were 92.6%, 96.7%, 85.1%, and 98.5%, respectively (Table 1). An "invalid" result was obtained in 0.7% (3/401) samples, indicating the sample processing control (SPC) did not meet the acceptable criteria; invalidity might be caused by inadequate processing of MRSA, specimen-associated inhibition of the real-time PCR assay, or air bubbles formed in the reaction tube.
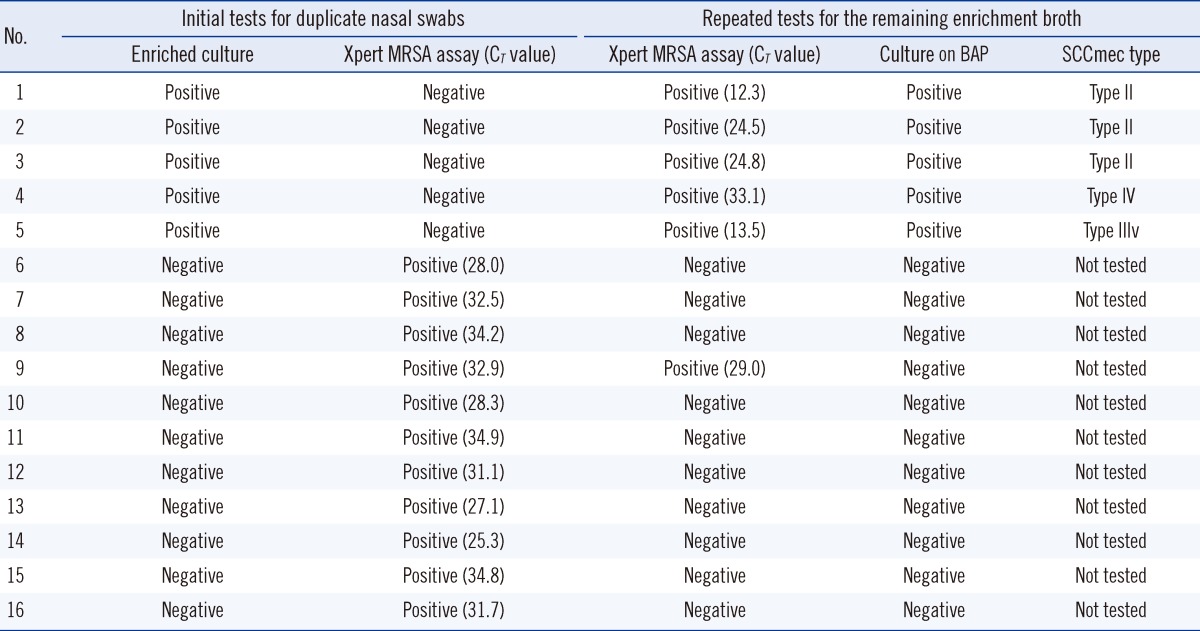
The discrepant results were observed in 16 samples (5 culture-positive, PCR-negative; 11 culture-negative, PCR-positive) (Table 2). For 5 samples showing culture-positive and PCR-negative results, the repeated Xpert MRSA assay using remaining TSB revealed all positive results and the CT values were variable (Table 2). For 11 samples showing culture-negative and PCR-positive results, the repeated Xpert MRSA assay using the remaining TSB revealed negative results for 10 samples. The mean CT value of 11 discordant samples with culture-negative and PCR-positive results was significantly higher than that of 63 concordant samples (31.0 vs. 24.6, respectively, P=0.001). However, the mean CT values between initial screening samples and follow-up screening samples were similar (25.9 vs. 25.1, respectively, P=0.597).
By re-culturing the remaining frozen TSB from 54 culture-positive patients, 52 MRSA isolates were obtained. For two TSB samples, the re-culturing failed. SCCmec type II was the most common (34/52); it was followed by SCCmec type IV (13/52), SCCmec type III variant (3/52), and SCCmec type IV variant (2/52). There was no difference in the detection rates of these SCCmec types among chromogenic agars and the X-pert MRSA assay.
In this study, three enriched-culture methods showed good overall performance for detecting MRSA. In particular, ChromID MRSA showed higher sensitivity, but its specificity was lower than Brilliance agar II. The detection sensitivity using ChromID MRSA increased up to 4.4% (from 95.6% at 24 hr to 100% at 48 hr) with extended incubation; inversely, the specificity decreased to 3.3% (from 95.8% at 24 hr to 92.5% at 48 hr), which corresponds to a previous report [10]. The results of Brilliance agar II (24 hr) were satisfactory at ≥90% with respect to sensitivity, specificity, PPV, and NPV for MRSA detection. ChromID MRSA (48 hr) showed the lowest specificity due to false-positive growth of Enterococcus spp. and methicillin-resistant coagulase-negative Staphylococci (MRCoNS). In contrast to the previous report where the MRCoNS and enterococci were the major species producing false-positive results on Brilliance agar I [10], Bacillus spp. was the major isolate in this study, and this problem was not observed with Brilliance agar II.
When the enriched culture was designated as a reference method, the sensitivity and specificity of the Xpert MRSA assay in this study were 92.6% and 96.7%, respectively, which were higher than those in the previous studies with a similar design. In one study for the comparison of Xpert MRSA assay with broth-enriched MRSA-Select chromogenic agar (Bio-Rad Life Science Group, Marnes-La-Coquette, France) with a prevalence of 22.1%, the sensitivity and specificity were 84% and 92%, respectively, for detecting MRSA from a nasal site [8]. In a multicenter study for the comparison of Xpert MRSA assay with broth-enriched CHROMagar (Becton, Dickinson and Company) with the prevalence of 5.2-44%, the sensitivity and specificity were 86.3% and 94.9%, respectively [6]. The analytical sensitivity (limit of detection, LoD) of the Xpert MRSA assay was about 50 colony-forming units (CFU) per swab, according to the manufacturer's instruction. It was also reported by Rossney et al. [8] that the LoD of the Xpert MRSA assay was 58 CFU per swab, and it was threefold lower but six-fold higher than those of direct and enrichment cultures, respectively. The higher specificity in this study might be due to the use of three kinds of media and a longer incubation time up to 48 hr. The mean CT value of the false-positive (culture-negative and PCR-positive) samples was significantly higher than that of concordant samples (31.0 vs. 24.6). These findings are in line with the previous studies showing that the discrepancies between the Xpert MRSA assay and culture method occurred exclusively when the bacterial density was low [9] and agree with the fact that the CT value in the Xpert MRSA assay correlates strongly with the bacterial density in the samples [9, 14]. The discordance from the five false-negative results might be partially explained by the cases with low bacterial density of MRSA in the nostrils or disparity in the specimens from two nostrils (Table 2). The reason for the one discrepancy of a sample yielding culture-negative and PCR-positive results, even in the repeated test using the remaining enrichment TSB, might be the fact that SCC elements lacking the functional mecA gene can be detected by the Xpert MRSA assay, because it does not specifically target the mecA gene [5]. Another explanation for the one culture-negative and PCR-positive sample is the limitation in the experimental design for S. aureus strains that may require anaerobic incubation [15].
There was no difference in the detection rates of SCCmec types among chromogenic agars and Xpert MRSA assay. The majority of the isolates were SCCmec type II that is known as HA-MRSA clone [12], followed by SCCmec type IV that is emerging as a CA-MRSA clone in Korea [16, 17].
In conclusion, the performance of the three chromogenic media combined with enriched culture and the Xpert MRSA assay was good (>92% of both sensitivity and specificity). Notably, the agreement rate between the Xpert MRSA assay and the enriched-culture methods was high (96%) in this study. The Xpert MRSA assay could give results comparable to culture methods within 1 hr. This early detection can help save 48-72 hr, and help in prevention and control of MRSA infection in health-care settings.
Acknowledgements
We thank Oxoid and Cepheid for providing chromogenic media and Xpert MRSA assay kits, respectively.
References
1. Yi J, Kim EC. Microbiological characteristics of methicillin-resistant Staphylococcus aureus. Korean J Clin Microbiol. 2010; 13:1–6.
2. Honda H, Krauss MJ, Coopersmith CM, Kollef MH, Richmond AM, Fraser VJ, et al. Staphylococcus aureus nasal colonization and subsequent infection in intensive care unit patients: does methicillin resistance matter? Infect Control Hosp Epidemiol. 2010; 31:584–591. PMID: 20426656.
3. Weber SG, Huang SS, Oriola S, Huskins WC, Noskin GA, Harriman K, et al. Legislative mandates for use of active surveillance cultures to screen for methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: position statement from the Joint SHEA and APIC Task Force. Infect Control Hosp Epidemiol. 2007; 28:249–260. PMID: 17326014.
4. Lee K, Kim MN, Kim JS, Hong HL, Kang JO, Shin JH, et al. Further increases in carbapenem-, amikacin-, and fluoroquinolone-resistant isolates of Acinetobacter spp. and P. aeruginosa in Korea: KONSAR study 2009. Yonsei Med J. 2011; 52:793–802. PMID: 21786445.
5. Arbefeville SS, Zhang K, Kroeger JS, Howard WJ, Diekema DJ, Richter SS. Prevalence and genetic relatedness of methicillin-susceptible Staphylococcus aureus isolates detected by the Xpert MRSA nasal assay. J Clin Microbiol. 2011; 49:2996–2999. PMID: 21677066.
6. Wolk DM, Picton E, Johnson D, Davis T, Pancholi P, Ginocchio CC, et al. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J Clin Microbiol. 2009; 47:758–764. PMID: 19129414.
7. Makgotlho PE, Kock MM, Hoosen A, Lekalakala R, Omar S, Dove M, et al. Molecular identification and genotyping of MRSA isolates. FEMS Immunol Med Microbiol. 2009; 57:104–115. PMID: 19712080.

8. Rossney AS, Herra CM, Brennan GI, Morgan PM, O'Connell B. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J Clin Microbiol. 2008; 46:3285–3290. PMID: 18685003.
9. Wolk DM, Marx JL, Dominguez L, Driscoll D, Schifman RB. Comparison of MRSASelect Agar, CHROMagar Methicillin-Resistant Staphylococcus aureus (MRSA) Medium, and Xpert MRSA PCR for detection of MRSA in Nares: diagnostic accuracy for surveillance samples with various bacterial densities. J Clin Microbiol. 2009; 47:3933–3936. PMID: 19828738.
10. Malhotra-Kumar S, Abrahantes JC, Sabiiti W, Lammens C, Vercauteren G, Ieven M, et al. Evaluation of chromogenic media for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010; 48:1040–1046. PMID: 20164268.
11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, M100-S20. Wayne, PA: CLSI;2010.
12. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005; 43:5026–5033. PMID: 16207957.
13. Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007; 51:3374–3377. PMID: 17576837.
14. Stenehjem E, Rimland D, Crispell EK, Stafford C, Gaynes R, Satola SW. Cepheid Xpert MRSA cycle threshold in discordant colonization results and a quantitative measure of nasal colonization burden. J Clin Microbiol. 2012; 50:2079–2081. PMID: 22442322.
15. Qian Q, Eichelberger K, Kirby JE. Rapid identification of Staphylococcus aureus in blood cultures by use of the direct tube coagulase test. J Clin Microbiol. 2007; 45:2267–2269. PMID: 17522280.
16. Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011; 66:1061–1069. PMID: 21393157.
17. Kim JS, Park JS, Song W, Kim HS, Cho HC, Lee KM. Panton-Valentine leukocidin positive Staphylococcus aureus isolated from blood in Korea. Korean J Lab Med. 2007; 27:286–291. PMID: 18094590.
Table 1
Comparison of Brilliance agar I, Brilliance agar II, ChromID MRSA, and Xpert MRSA assay for screening of MRSA from 401 nasal swabs
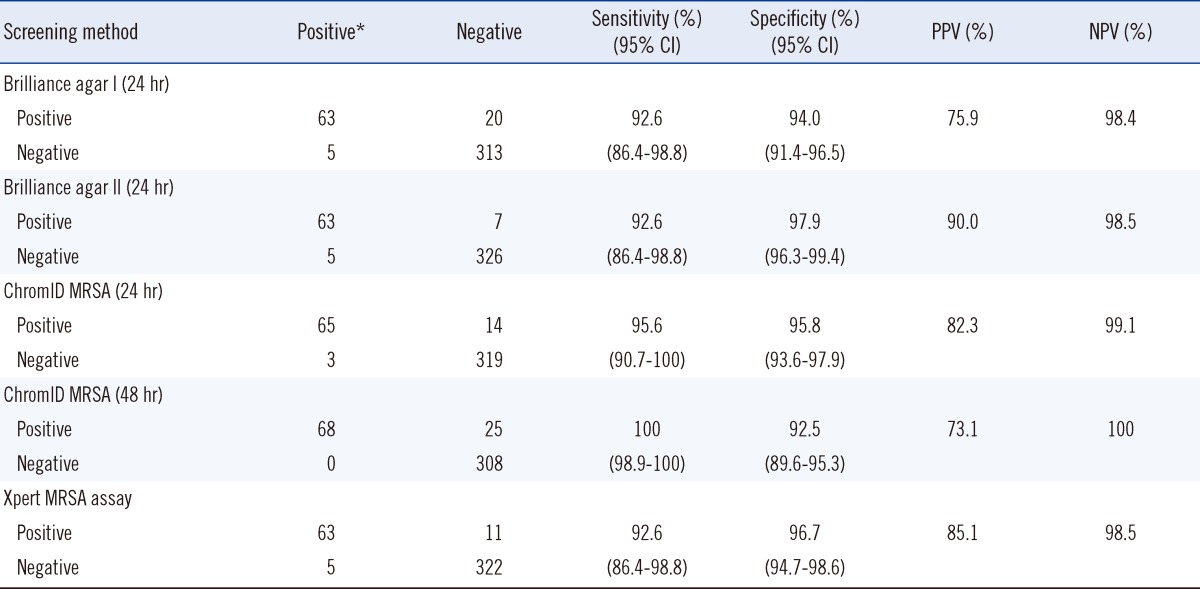
*A true positive culture was determined if MRSA was detected in one or more of the three culture media tested, and it was used as a reference culture result.
Abbreviations: MRSA, Methicillin-resistant Staphylococcus aureus; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download