Abstract
Background
We reviewed patients with multiple myeloma (MM) in order to assess the incidence of genetic abnormalities and their associations with clinical parameters, risk groups, and prognosis.
Methods
A total of 130 patients with MM were enrolled. The incidences of genetic abnormalities were determined in all patients. The relationships of the genetic abnormalities and clinical parameters were investigated. In addition, a survival analysis was performed.
Results
Abnormal karyotypes were detected in 42.3% (N=55) of the patients, and this was increased to 63.1% (N=82) after including the results determined with interphase FISH. Hypodiploidy was observed in 7.7% (N=10) of the patients, and all were included in the group with complex karyotypes (30.8%, N=40). The 14q32 rearrangements were detected in 29.2% (N=38) of the patients, and these most commonly included t(11;14), which was followed by t(4;14) and t(14;16) (16.2%, 11.5%, and 0.8%, respectively). Abnormal karyotypes and complex karyotypes were associated with disease progression markers, including low hemoglobin levels, low platelet counts, high plasma cell burden, high β2-microglobulin, and high international staging system stages. A high free light chain (FLC) ratio and FLC difference were associated with abnormal karyotypes, complex karyotypes, and higher plasma cell burden. Hypodiploidy and low platelet counts were significant independent prognostic factors and were more important in patient outcome than any single abnormality.
Multiple myeloma (MM) is characterized by the clonal proliferation of plasma cells and their accumulation within the bone marrow (BM) [1]. Over the past few years, one or more new agents have been incorporated into the treatment paradigm of patients with MM, increasing survival rates of MM patients [2]. The prognoses of patients with MM are highly variable. While a portion of patients survive over a decade after their diagnosis, a small subset at the other end of the spectrum exhibits a highly aggressive course [3]. Prognostic factors for MM have been suggested, and the International Staging System (ISS) is most widely applied. It stratifies patients by their albumin and β2-microglobulin (β2M) levels [4]. The presence of genetic abnormalities in plasma cells has been considered to be an important prognostic factor, and several genetic abnormalities have been extensively investigated during the past decade [5-7]. Although many genetic abnormalities are amenable to detection by conventional cytogenetics, their detection is impaired by the relatively low rate of proliferation and, consequently, the low proportion of cells in metaphase [3]. Some aberrations are cryptic and cannot be detected by conventional cytogenetics. These limitations can be overcome in part by interphase FISH [8, 9].
We retrospectively reviewed a database of patients with MM from a single institute in Korea in order to assess the incidence of genetic abnormalities and their relationships with clinical parameters and prognosis.
A total of 130 patients with de novo MM were enrolled. The demographic, laboratory, and clinical parameters of the patients are listed in Table 1. Among them, 47 (36.2%) of the patients were administered novel agents, including bortezomib or thalidomide, and 39 patients underwent hematopoietic stem cell transplantation (HSCT) [10]. The study was performed according to the Declaration of Helsinki and approval for this study was obtained from the Institutional Review Board of St. Mary's Hosptial, The Catholic University of Korea (KC09EISI0393).
Chromosomal analyses were performed by diagnosis of short-term cultures of BM specimens and standard conventional cytogenetic protocols. At least 20 metaphases were analyzed in each case, and the clonal abnormalities were classified according to the 2009 international system for human cytogenetic nomenclature (ISCN) guidelines [11].
All samples were investigated with interphase FISH with the following probes: LSI IGH Dual Color, Break Apart Rearrangement Probe, LSI IGH/CCND1, IGH/FGFR3, IGH/MAF dual color, dual fusion translocation probe, LSI TP53 (17p13.1)/CEP 17 probe, and LSI 13 (RB1) 13q14 probe (Vysis Inc., Abbott Laboratories, Abbott Park, IL, USA). BM cells from routine cytogenetic preparations were used. The results were considered abnormal if the percentage of nuclei with abnormal signals exceeded the normal reference ranges. The following cut-off levels for positive results were set according to the European Myeloma Network recommendations: 10% for the IGH break apart probe and for both translocations and 20% for deletion 13q14 (del(13q)) and for deletion 17p13 (del(17p)) [12].
FLC measurements were determined with a commercial kit (FREELITE, The Binding Site Group Ltd., Birmingham, UK) on a Toshiba 200 FR Neo analyzer (Toshiba Medical Systems Corporation, Tokyo, Japan). The ratio of involved to uninvolved FLC (FLC ratio) was calculated, and 277 was used as a cutoff value. The absolute difference between the involved and uninvolved FLC (FLC diff) was also calculated, and 185 was adopted as a cutoff value, as described previously [13].
The associations between genetic abnormalities and clinical parameters were analyzed by Pearson's Chi-square tests and Fisher's exact tests. The Kaplan-Meier method was used to estimate the probability of survival. The statistical significance of the factors that were associated with overall survival (OS) was investigated by univariate and multivariable Cox proportional hazards regression models. Hazard ratios (HR) and their 95% confidence intervals (CI) were computed. Statistical significance was defined as P values that were less than 0.05. All of the statistical analyses were performed with MedCalc software 9.0 (MedCalc Software, Ostend, Belgium).
Abnormal karyotypes were detected in 42.3% (55/130) of the patients. Patients with 1-2 karyotypic abnormalities made up 11.5% (N=15) of the patients, while a complex karyotype was observed in 30.8% (N=40) of the patients. Thus, 72.7% of all abnormal karyotypes corresponded to a complex karyotype. Hypodiploidy was observed in 7.7% (N=10) of all patients, and all of them were included in the complex-karyotype group. Hyperdiploidy was detected in 16.9% (N=22) of the patients with conventional cytogenetics. After including the results from the FISH analysis, 82 patients (63.1%) exhibited genetic abnormalities.
A 14q32 rearrangement was detected in 29.2% (N=38) of the patients, and these most commonly included t(11;14), which was followed by t(4;14) and t(14;16) (16.2%, 11.5%, and 0.8%, respectively). del(13q) was a common genetic abnormality, and it had an incidence of 26.9% (35/130). Sixty percent (9/15) of the patients had t(4;14) and del(13q), and this incidence was significantly higher than patients without t(4;14) (22.6%, P=0.002). A Gain of 1q21 (1q+) was found in 16.9% (N=22) of the patients, and all of them were included in the complex-karyotype group. The incidence of del(17p) was 10.8% (N=14), and 78.6% of them (11/14) also contained del(13q).
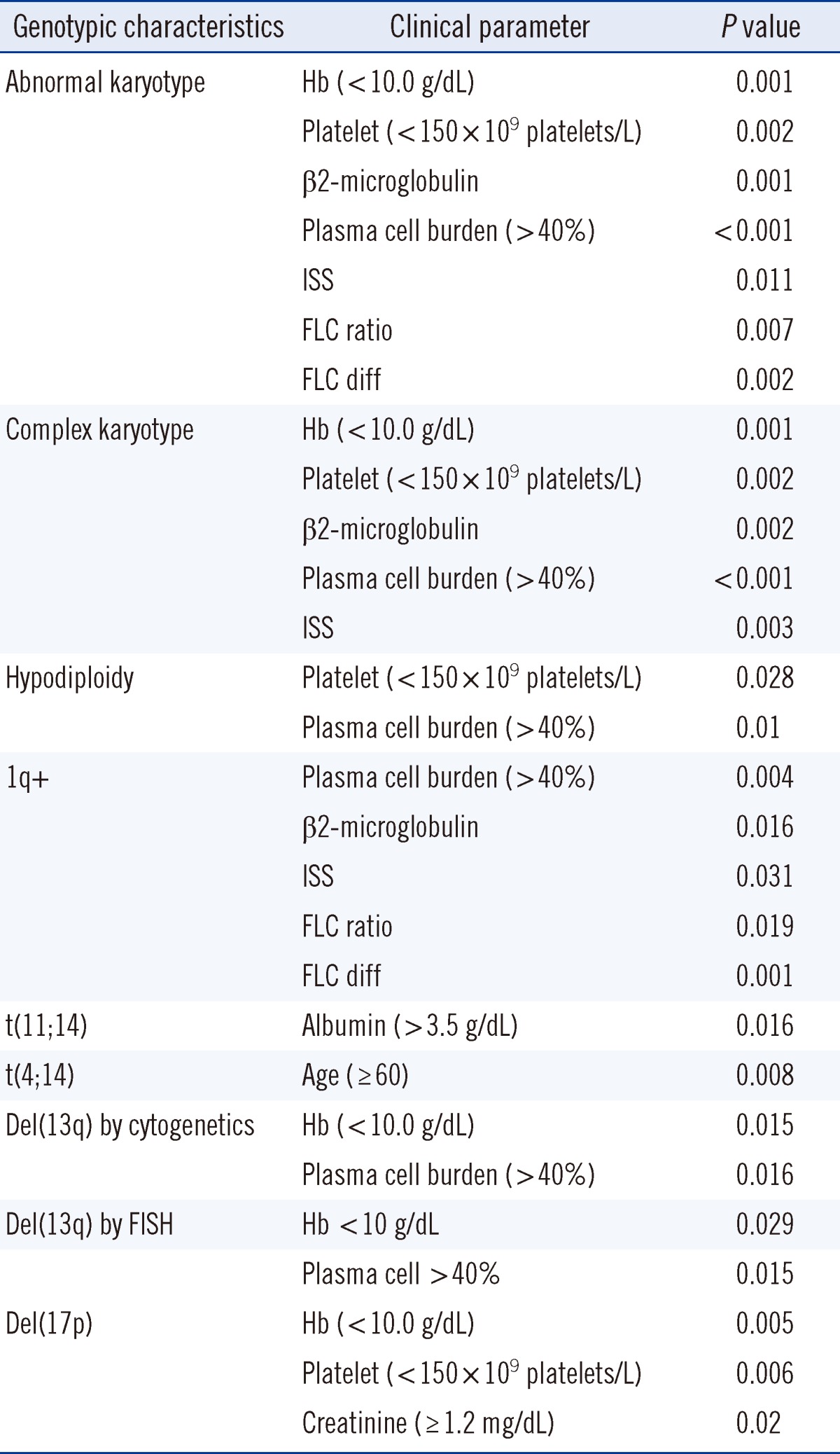
The associations of genetic abnormalities and clinical and laboratory parameters are listed in Table 2. Low hemoglobin (Hb<10 g/dL) levels were associated with abnormal karyotypes, complex karyotypes, hyperdiploidy, del(13q), and del(17p). Low platelet counts (<150×109/L) were frequently found in patients with abnormal karyotypes, complex karyotypes, hypodiploidy, or del (17p). High plasma cell burdens (≥40%) were associated with abnormal karyotypes, complex karyotypes, hypodiploidy, del (13q), and 1q+. High β2M levels (≥5.5 mg/dL) and high ISS stages were frequently found in patients with abnormal karyotypes and complex karyotypes. t(11;14) was associated with higher albumin levels (albumin>3.5 g/dL). t(4;14) was more common in patients who were over 60 yr of age, and del (17p) was associated with decreased renal function, as indicated by high creatinine (Cr) levels (Cr ≥1.2 mg/dL). A high FLC ratio and FLC diff were associated with high plasma cell burdens (P=0.007 and P=0.002, respectively), abnormal karyotypes (P=0.020 and P=0.020, respectively), and 1q+ (P=0.019 and P=0.001, respectively). High FLC ratios were associated with 14q32 rearrangements (P=0.047). The median (range) of the FLC ratios of patients with 14q32 rearrangements and negative controls were 158.7 (1.0-15,325.0) and 55.4 (0.6-2,807.5), respectively.
The median survivals in months according to genetic abnormalities are listed in Table S1 (supplemental data; available in online). The median follow-up duration was 60 months (minimum-maximum, 6-575 months). A total of 25 of the 130 patients (19.2%) died during the study. Novel agents, including bortezomib or thalidomide, were administered to 47 (36.2%) of the patients, and 39 (30.0%) patients underwent HSCT. Their OS rates were comparable to patients who did not receive novel agents or HSCT. The median OS of patients with the complex karyotype was 46 months (95% CI, 28.9-63.1), which was less than those for patients with 1-2 abnormalities and those with a normal karyotype (Fig. 1A, P=0.004). The median OS of patients with a complex karyotype with hypodiploidy was 15 months (95% CI, 0-37.2), which was significantly less than the 53 months (95% CI, 42.4-63.6) for patients with complex karyotypes without hypodiploidy (Fig. 1B, P<0.001). Hyperdiploidy, del(13q), 1q+, 14q32 rearrangement, t(11;14), t(4;14), and del(17p) affected the OS, but none of them exhibited statistical significance.
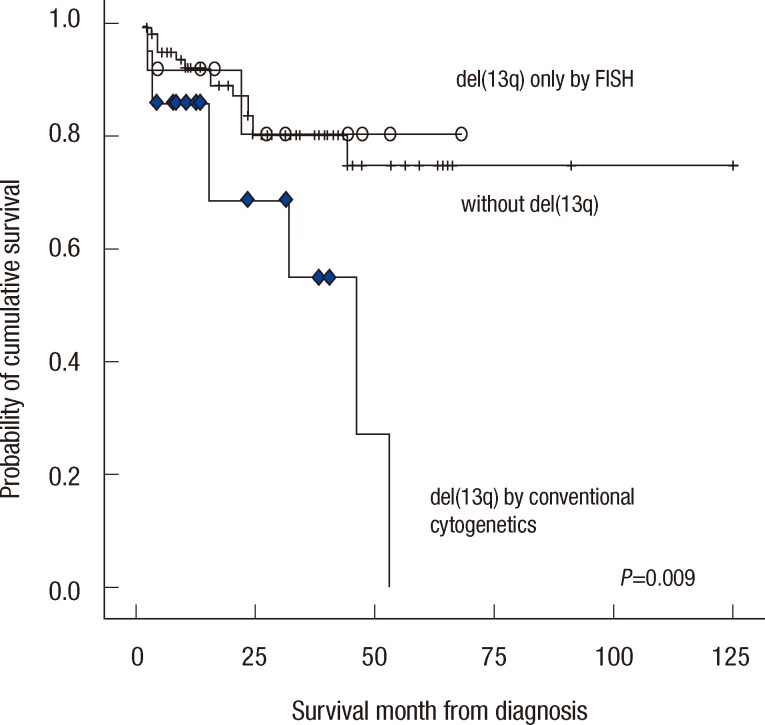
The OS of patients with del(13q) that was detected with conventional cytogenetics was 46.0 (95% CI, 25.1-66.9), which was compared to the median that was not reached for the patients with del(13q) that was detected by FISH and negative control (Fig. 2). Statistical significance was observed between the findings with conventional cytogenetics and negative controls (P=0.009). The OS of patients with del(13q) that was detected by FISH did not exhibit a significant difference compared to negative controls (P=0.872). Therefore, cytogenetic del(13q) was more associated with poor prognosis than the others (P=0.008).
The ISS stage, several genetic variables, and other clinical parameters were assessed with a univariate analysis for survival in order to identify significant prognostic effects. The prognostic variables for the OS multivariable model included age, ISS, Hb levels, platelet counts, abnormal karyotype, complex karyotype, and hypodiploidy. Platelet counts (HR, 3.740; 95% CI, 0.989-14.137, P=0.035) and hypodiploidy (HR, 9.614; 95% CI, 2.251-41.060, P=0.008) were significant independent prognostic factors for OS based on the final multivariable model that was obtained by the stepwise selection of variables.
Genetic abnormalities in MM, which are typically complex and represent a hallmark of the disease, involve many chromosomes that are altered both in number and structure. The standard diagnostic workups of these patients now include conventional cytogenetics to detect hypodiploidy and del(13q) and interphase FISH to detect 14q32 rearrangements, including t(11;14), t(4;14), t(14;16), hyperdiploidy, and del(17p) [14]. In this study, we demonstrated conventional cytogenetic abnormalities in 42.3% of the MM patients, which was increased to 63.1% of the patients after including the results that were determined by interphase FISH. Conventional cytogenetics detected abnormal chromosomes in as few as 26-40% of the cases due to the low proliferative activity of MM [15, 16], whereas interphase FISH in combination with cytoplasmic immunoglobulin staining or immunomagnetic separations of the plasma cells enable the detection of specific abnormalities in up to 86-98% of the patients [17-20]. The detection rates depended on the number of FISH probes, the plasma cell loads in the sample, and the selection procedures of the plasma cells. For del(13q), the sensitivity of conventional cytogenetics is lower compared to that of interphase FISH because of the slow division of neoplastic plasma cells, whereas FISH is independent of plasma cell division and has a higher yield of detecting genetic abnormalities [21, 22]. Recently, the European Myeloma Network has reported several recommendations for establishing standards for FISH analyses of newly diagnosed cases of MM. They suggested that the essential abnormalities are t(4;14), t(14;16), and del(17p), and at least 100 purified or specifically identified plasma cells should be scored [12].
The number of chromosomes has prognostic significance. Hypodiploidy is associated with poorer OS, while hyperdiploidy is better. Our data demonstrated that all patients with hypodiploidy were included in the complex-karyotype group. Interestingly, the prognoses of patients with hypodiploidy were worse than those with complex karyotypes without hypodiploidy. The incidence of hyperdiploidy was 16.9% with conventional cytogenetics, and the majority (9/22) of them revealed a low proportion of abnormal karyotypes (<25%). This indicated that hyperdiploidy can be more effectively detected by interphase FISH than by conventional cytogenetics.
The 14q32 rearrangements are widely recognized prognostic parameters. However, t(11;14) is prognostically neutral or associated with a better prognosis, and t(4;14), t(14;16), and t(14;20) are associated with poor prognoses. Although the limited number of patients in this study restricted the clarification of the prognostic significance, t(11;14) was associated with higher albumin levels, which may be related to mild clinical manifestation. t(4;14) was more frequently found in old patients, and it was associated with del(13q) that was detected with conventional cytogenetics and that is a well-known intermediate risk factor [23].
Secondary genetic aberrations, including the translocation of MYC, del(13q), deletions and/or amplifications of chromosome 1, and del(17p), have been found with disease progression [24]. We detected 3% with MYC rearrangements, 26.9% with del (13q), 16.9% with 1q+, and 10.8% with del(17p). It is notable that 78.6% of cases with del(17p) also contained del(13q), and all of the cases with 1q+ were included in the complex-karyotype group. These genetic aberrations were associated with disease progression markers, including low Hb levels, low platelet counts, high Cr levels, and high plasma cell burden.
A number of associations of genetic abnormalities and clinical parameters have been reported. The t(11;14)(q13;q32) is related to lymphoplasmacytic morphology, CD20 expression, and specific subtypes, including IgM, IgE, and nonsecretory plasma cell myeloma [25, 26]. t(4;14) is related to patient ages over 60, IgA-type plasma cell myeloma, and genetic abnormalities, including del(13q) and hypoploidy [26, 27]. del(17p) is frequently found in patients with plasma cell leukemia, the involvement of the central nervous system, and poor prognoses [26].
Kumar et al. [13] have hypothesized that 14q32 rearrangements lead to the unbalanced production of light chains and more extreme abnormalities of FLC. They have demonstrated that an abnormal FLC diff and FLC ratio are frequently detected in patients with 14q32 rearrangements and are associated with poor prognosis, especially in patients with t(14;16). We demonstrated that the FLC ratios were higher in patients with 14q32 rearrangements. In addition, the FLC ratio and FLC diff were associated with 1q+ and plasma cell burden. These findings support the idea that an increase in the FLC is due to the cumulative effect of several events, such as genetic aberrations, including 14q32 rearrangements and 1q+ and plasma cell proliferative activity, rather than any single abnormality.
The risks of stratification and treatment strategies are being updated continuously. With the advances of FISH studies and with the increasing number of probes used, it has become clear that nearly all patients with MM have one or more abnormalities. The presence of trisomies that are detected by FISH in patients with high-risk cytogenetic abnormalities ameliorates the usual adverse impact of t(4;14), t(14;16), t(14;20), or del(17p) [20]. With the use of frontline bortezomib-based regimens, the poor prognostic impact of t(4;14) has been overcome [27]. In addition, bortezomib may effectively surmount the adverse impact of chromosomal 13 abnormalities. In 2012, t(4;14), karyotypic del (13q), or hypodiploidy were updated as intermediate-risk diseases instead of their designation as high risk in 2011 [28]. This study demonstrated that hypodiploidy and low platelet counts remained significantly independent prognostic factors, showing that the cumulative effects of genetic abnormalities and clinical parameters are more important for patient outcome than any single abnormality. Thus, more data accumulation of long-term follow-up studies is necessary for adequate risk stratification.
Acknowledgements
This study was supported by a grant of the Korea Health Technology R$D Project, Ministry of Health $ Welfare, Republic of Korea (A120175).
References
1. Boyd KD, Ross FM, Chiecchio L, Dagrada GP, Konn ZJ, Tapper WJ, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012; 26:349–355. PMID: 21836613.

2. Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008; 111:2516–2520. PMID: 17975015.

3. Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood. 2007; 109:3489–3495. PMID: 17209057.
4. Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Bladé J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005; 23:3412–3420. PMID: 15809451.

5. Wuilleme S, Robillard N, Lodé L, Magrangeas F, Beris H, Harousseau JL, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005; 19:275–278. PMID: 15538401.

6. Cremer FW, Bila J, Buck I, Kartal M, Hose D, Ittrich C, et al. Delineation of distinct subgroups of multiple myeloma and a model for clonal evolution based on interphase cytogenetics. Genes Chromosomes Cancer. 2005; 44:194–203. PMID: 16001433.

7. Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. PMID: 19798094.

8. Drach J, Schuster J, Nowotny H, Angerler J, Rosenthal F, Fiegl M, et al. Multiple myeloma: high incidence of chromosomal aneuploidy as detected by interphase fluorescence in situ hybridization. Cancer Res. 1995; 55:3854–3859. PMID: 7641204.
9. Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed but correlate with natural history, immunological features and clinical presentation. Blood. 2002; 99:2185–2191. PMID: 11877296.

10. Min CK, Lee MJ, Eom KS, Lee S, Lee JW, Min WS, et al. Bortezomib in combination with conventional chemotherapeutic agents for multiple myeloma compared with bortezomib alone. Jpn J Clin Oncol. 2007; 37:961–968. PMID: 18156171.

11. Shaffer LG, Slovak ML, editors. ISCN 2009: an international system for human cytogenetic nomenclature. Basel: Karger;2009.
12. Ross FM, Avet-Loiseau H, Ameye G, Gutiérrez NC, Liebisch P, O'Connor S, et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012; 97:1272–1277. PMID: 22371180.

13. Kumar S, Zhang L, Dispenzieri A, Van Wier S, Katzmann JA, Snyder M, et al. Relationship between elevated immunoglobulin free light chain and the presence of IgH translocations in multiple myeloma. Leukemia. 2010; 24:1498–1505. PMID: 20520636.

14. Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol. 2012; 87:78–88. PMID: 22180161.

15. Kröger N, Schilling G, Einsele H, Liebisch P, Shimoni A, Nagler A, et al. Deletion of chromosome band 13q14 as detected by fluorescence in situ hybridization is a prognostic factor in patients with multiple myeloma who are receiving allogeneic dose-reduced stem cell transplantation. Blood. 2004; 103:4056–4061. PMID: 14982868.

16. Christensen JH, Abildgaard N, Plesner T, Nibe A, Nielsen O, Sørensen AG, et al. Interphase fluorescence in situ hybridization in multiple myeloma and monoclonal gammopathy of undetermined significance without and with positive plasma cell identification: analysis of 192 cases from the Region of Southern Denmark. Cancer Genet Cytogenet. 2007; 174:89–99. PMID: 17452249.

17. Dewald GW, Therneau T, Larson D, Lee YK, Fink S, Smoley S, et al. Relationship of patient survival and chromosome anomalies detected in metaphase and/or interphase cells at diagnosis of myeloma. Blood. 2005; 106:3553–3558. PMID: 16030187.

18. Schmidt-Wolf IG, Glasmacher A, Hahn-Ast C, Jüttner A, Schnurr T, Cremer F, et al. Chromosomal aberrations in 130 patients with multiple myeloma studied by interphase FISH: diagnostic and prognostic relevance. Cancer Genet Cytogenet. 2006; 167:20–25. PMID: 16682281.

19. Ross FM, Ibrahim AH, Vilain-Holmes A, Winfield MO, Chiecchio L, Protheroe RK, et al. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia. 2005; 19:1634–1642. PMID: 15990862.

20. Kumar S, Fonseca R, Ketterling RP, Dispenzieri A, Lacy MQ, Gertz MA, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012; 119:2100–2105. PMID: 22234687.

21. Kapoor P, Fonseca R, Rajkumar SV, Sinha S, Gertz MA, Stewart AK, et al. Evidence for cytogenetic and fluorescence in situ hybridization risk stratification of newly diagnosed multiple myeloma in the era of novel therapie. Mayo Clin Proc. 2010; 85:532–537. PMID: 20511484.
22. Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Cheis M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004; 64:1546–1558. PMID: 14989251.
23. Kumar SK, Mikhael JR, Buadi FK, Dingli D, Dispenzieri A, Fonseca R, et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines. Mayo Clin Proc. 2009; 84:1095–1110. PMID: 19955246.

24. Kapoor P, Rajkumar SV. Update on risk stratification and treatment of newly diagnosed multiple myeloma. Int J Hematol. 2011; 94:310–320. PMID: 22005834.

25. Avet-Loiseau H, Garand R, Lodé L, Harousseau J, Bataille R. Intergroups Francophone Myélome. Translocation t(11;14)(q13;q32) is the hallmark of IgM, IgE, and nonsecretory multiple myeloma variants. Blood. 2003; 101:1570–1571. PMID: 12393502.

26. Stewart AK, Bergsagel PL, Greipp PR, Dispenzieri A, Gertz MA, Hayman SR, et al. A practical guide to defining high-risk myeloma for clinical trials, patient counseling and choice of therapy. Leukemia. 2007; 21:529–534. PMID: 17230230.

27. Butler C, Wolff DJ, Kang Y, Stuart RK, Costa LJ. Association of age with fluorescence in situ hybridization abnormalities in multiple myeloma reveals higher rate of IGH translocations among older patients. Leuk Lymphoma. 2012; 53:2444–2448. PMID: 22574971.
28. San Miguel JF, Schlag R, Khuaqeva NK, Dimopoulos MA, Shpilberg O, Kropff M, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008; 359:906–917. PMID: 18753647.

29. Rajkumar SV. Multiple myeloma: 2011 update on diagnosis, risk-stratification, and management. Am J Hematol. 2011; 86:57–65. PMID: 21181954.

Fig. 1
Survival curves for patients with different genetic abnormalities. Patients with complex karyotypes (rhombus) exhibited unfavorable prognoses compared to normal patients (line) and those with 1-2 abnormalities (square) (A). Patients with hypoploidy (rhombus) exhibited unfavorable prognoses compared to patients without genetic abnormalities (line) (B).
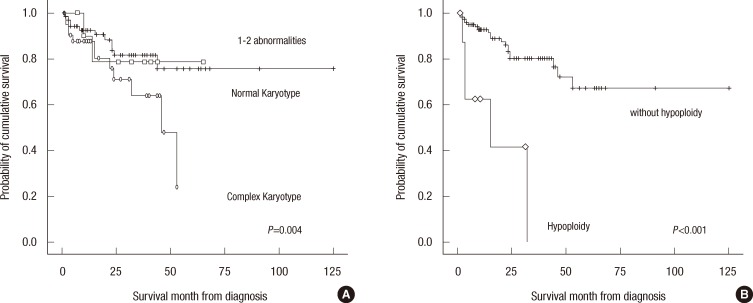
Fig. 2
Patients with 13q deletions detected by conventional cytogenetics (rhombus) exhibited unfavorable prognoses compared to patients with del(13q) detected only by FISH (circle) or without del(13q) (line).




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download