Abstract
Clinical microbiology has always been a slowly evolving and conservative science. The sub-field of bacteriology has been and still is dominated for over a century by culture-based technologies. The integration of serological and molecular methodologies during the seventies and eighties of the previous century took place relatively slowly and in a cumbersome fashion. When nucleic acid amplification technologies became available in the early nineties, the predicted "revolution" was again slow but in the end a real paradigm shift did take place. Several of the culture-based technologies were successfully replaced by tests aimed at nucleic acid detection. More recently a second revolution occurred. Mass spectrometry was introduced and broadly accepted as a new diagnostic gold standard for microbial species identification. Apparently, the diagnostic landscape is changing, albeit slowly, and the combination of newly identified infectious etiologies and the availability of innovative technologies has now opened new avenues for modernizing clinical microbiology. However, the improvement of microbial antibiotic susceptibility testing is still lagging behind. In this review we aim to sketch the most recent developments in laboratory-based clinical bacteriology and to provide an overview of emerging novel diagnostic approaches.
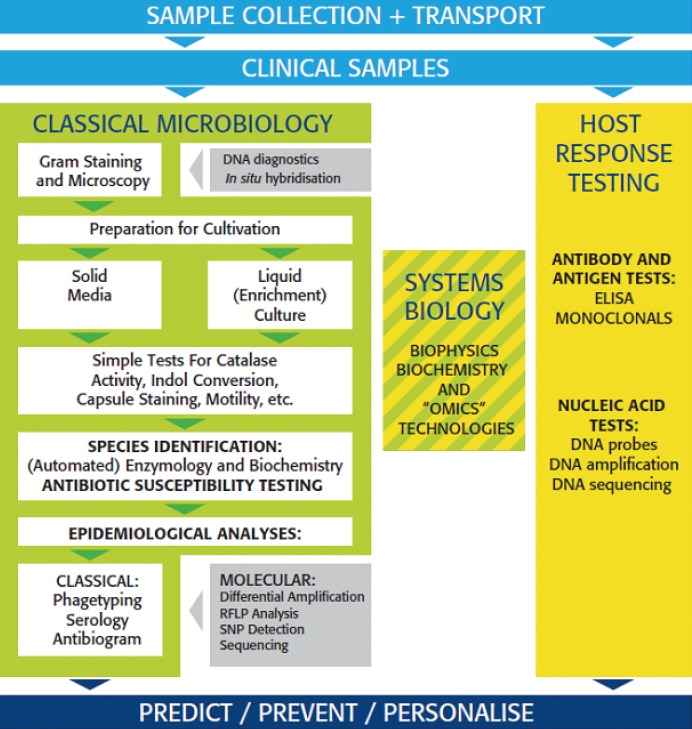
Clinical microbiology and bacteriology in particular has always been a slowly evolving science. Technologies developed in the 19th century still play a central role in routine diagnostics and subsequent therapeutic responses. Classical growth-based microbiology similar to that employed by Pasteur and Koch is not only used today for microbial detection and identification, it also provides the technological basis for much of the antimicrobial susceptibility testing (AST) that is performed. In addition, even older technologies, including a form of light microscopy that would be recognizable by Van Leeuwenhoek himself, continue to be commonplace. Fig. 1 summarizes today's workflow in this conservative environment. In the bacteriology laboratory, incoming samples are usually subjected to Gram-staining and microscopy after which the same materials are prepared for cultivation on solid or in liquid growth media. After incubation for a sufficient duration at an appropriate temperature in a suitable atmosphere, microbial growth is evaluated. Samples without microbial growth are discarded whereas positive samples are evaluated further including more detailed strain characterization (often initiated by a second and time-consuming round of purifying cultivation) using simple and inexpensive testing (motility assessment, catalase testing etc.). This leads to (partial) species assessment, which may be completed using manual or automated biochemical testing and extended enzymology [1]. Next or sometimes even simultaneously, the bacterial isolates can be further characterized with respect to their antimicrobial susceptibility profiles and their epidemiological characteristics. For these studies, many classical and some automated basic microbiology methods have been made available [2, 3]. This overall picture essentially describes the core activities of a clinical microbiology laboratory as it has functioned for the past century.
More recently, several waves of mostly technologically oriented innovation have been observed. With the advent of immunological methods, measurements of host reactivity to infection were facilitated. Definition of antibody specificity and levels using ELISA, co-determined by the exquisite specificity of monoclonal antibodies, became an accepted diagnostic standard for the detection of many microbial infections [4]. Similar serological tests have also been used for epidemiological characterization of microorganisms [5]. After the establishment of immunodiagnostics the development of molecular-based testing was not far behind. Although primarily developed in research oriented laboratories, molecular technology has acquired an important place in clinical microbiology. The direct detection of nucleic acids specific for certain microbial species has revolutionized, for instance, the detection of the sexually transmitted bacterial species Chlamydia trachomatis and Neisseria gonorrhoeae [6]. The use of DNA probes and nucleic acid amplification became well accepted although the analytical specificity and sensitivity of some of these tests may still require optimization [7]. Today biophysical technology is entering the clinical microbiology arena (e.g. matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) [8]) and this will allow for further automation of laboratory procedures.
Hence, over the past two decades clinical microbiology laboratories have been genuinely transformed. Direct host testing became more integrated and molecular and biophysical diagnostic technologies were successfully introduced. Over the coming years, the sequential or combined introduction of aspects of the other "omics" technologies (genomics, transcriptomics, proteomics, glycomics, lipidomics etc; for a recent review, see [9]) will further enhance the implementation of real-time rational therapies and the improvement of microbiological surveillance [10, 11]. This approach will, in turn, lead to prediction, prevention and personalization of the infectious risk assessment and, hopefully, more efficacious treatment of infectious diseases.
This short review aims to describe and position some of the recent technological advances and breakthroughs and to identify current shortcomings in the functioning of the clinical bacteriology laboratory.
Clinical specimens and their management are key quality determinants in clinical microbiology. From the moment a specimen is collected, timing becomes critical for microbial viability i.e., the longer specimens are held under bacterial growth-limiting conditions, the smaller the chance of recovering microorganisms by growth-based methods. This, in turn, can lead to drastic performance changes in test sensitivity and specificity as well. Strictly anaerobic organisms, for instance, may ultimately be overgrown by even small numbers of the conditional anaerobic or fully aerobic species. This means that speed of transportation is an important quality parameter in clinical microbiology [12]. Unfortunately the clinical laboratory often cannot stringently control this aspect of quality assurance other than outright rejection of poorly handled clinical materials. Policies to improve awareness are very important in this respect. From a research and development point of view there is an obvious need for improvement of transport media when organism viability is essential. Since some bacterial species are considered to be uncultivable on the presently used artificial growth media, there is also an obvious need for designing new media or alternative culture formats. When diagnostics involve the detection of DNA or other cellular components, a good lysis buffer containing compound-specific stabilizers (e.g. DNase or protease inhibitors) is equally important [13]. The diverse nature of clinical specimens including the biological diversity normally encountered even in different samples of the same clinical material renders adequate detection and quantification a complex and challenging task. Equipment that would facilitate the parallel purification of host cells, bacteria, nucleic acids and/or viruses and proteins or other sub-cellular elements from diverse clinical materials would certainly be both clinically and commercially successful [14]. Obviously, the development of methods that utilize direct enumeration of potentially pathogenic microorganisms using stabilized nucleic acids in samples are less susceptible to transport stringency.
Culture has been the mainstay of clinical microbiology for the past century and will likely remain so for decades to come despite accelerating technological development and the introduction of novel diagnostic procedures. However, there remains an absolute need for significant quantities of living organisms - not as much for detection and identification of microbial species, since strong alternative technologies are being introduced including MALDI-TOF MS, but more specifically for AST. The ability to document the killing or stationary effect (or lack thereof) of antibiotics on intact, living cells has not yet found a suitable growth-free alternative although advances toward this goal are currently under development [15]. Bacterial killing by antibiotics is usually monitored by the visible changes in bacterial density in liquid or solid growth media in the absence or presence of strategic concentrations of those antibiotics with clinical relevance. The read-out technology may change in the coming years but straightforward live-dead monitoring is still at the core of AST until bacterial death can be reliably and reproducibly measured in single cells or small numbers of cells in a culture-free environment. To date, most applications require between 104 and 105 microbial cells per assay.
However, classical culture is also being challenged and optimized continuously. The standard Petri dish format is faced with competing films, fibers and nano-porous carrier materials in which the actual "growth containers" may have internal volumes in the nano-liter range. This enables development of contained growth and/or micro-monitoring on alumina oxide chips for example [16, 17]. Solid agar media can be loaded with chromogenic compounds that are specifically metabolized by a certain bacterial species only [18]. This facilitates a direct on-the-plate, visual species identification that hastens detailed reporting of culture results. When antibiotics are included in the chromogenic or standard agar-based media, it might provide presumptive species identification but also limited resistance screening [19, 20]. It must be emphasized that AST based upon growth inhibition on solid media is still immensely popular. Disk diffusion testing and/or EtestR are still used as a primary means of AST in many laboratories [21]. The automation of solid media inoculation is becoming extremely important for large sample volume laboratories. A number of systems are currently marketed for that purpose including mechanisms for automated incubation and overall management of actively growing cultures. Such machines usually harbor cameras for real time archiving of (growing) cultures, demonstrating that detection and morphological characterization of (micro-)colonies may potentially speed up time to results for culture-based technology, certainly if growth could be monitored from a distance (e.g. via an internet connection with the incubator's data acquisition station) [22]. It has to be noted that such requirements for small versus big laboratories or laboratories in developed versus developing countries may be hugely different.
Liquid culture is continuously optimized as well - not only by changing the medium composition but also by progressive automation and the use of increasingly sophisticated growth sensors. Special compounds, such as resins that specifically capture and inactivate antibiotics present in patient samples, are added to reduce the time to positivity [23]. AST in liquid phase is easily automated and a number of systems are available for the clinical microbiology laboratory including the Becton Dickinson Phoenix, the Siemens Micoscan WalkAway or the bioMérieux VITEK2™ systems, which are the three best-known examples [24].
It has to be noted that there are no studies that unambiguously demonstrate the added value of "rapidity adaptations" of classical microbiology methods [25, 26]. This has to be realized by those involved in microbiological R&D and the focus should clearly remain on the acceleration of these improvements. Obviously, alternative approaches rather than classical methodology may then be obligatory.
Clinical microbiology is evolving in a speed that is much higher than it used to be in the previous century. As a consequence, the availability of experienced laboratory personnel is becoming a critical issue and although properly trained technicians are rare, there is a clear need to adapt their training to meet the diagnostic expectations. Current developments in clinical microbiology, in one way or another, target laboratory automation, high level information generation and reducing the overall time to results of detection, identification, and AST of bacteria, yeasts and molds in general. Secondary trends are related to connectivity (between automated systems, between systems and the laboratory or hospital information systems (LIS, HIS) and between the clinical microbiologist and the physician), cost effectiveness, quality services and clinical information content (how to deliver the most useful microbiological information as quickly as possible to clinicians). As technological innovations, imaging, mass spectrometry (MS) and sequencing are at the core of diagnostic R&D and so too are possible improvements in the speed, accuracy, accessibility and clinical correlation of AST results. Below, a selection of such developments will be addressed in more detail.
Nucleic acid testing was introduced in the 1980's. Plasmid purification and (in situ) nucleic acid probe-mediated detection, identification and characterization were the first technologies developed [27]. Neither of these was broadly accepted by the clinical community. The same happened to restriction fragment length polymorphism (RFLP) analysis and a few other even more obscure molecular test formats [28]. However, epidemiological typing using pulsed field gel electrophoresis (PFGE) of DNA macro-restriction fragments became an undisputed gold standard in many laboratories [29] and public health laboratories in particular. And of course sequencing (see separate section below) and PCR became much more appreciated over the past decade. PCR by itself has revolutionized the science of clinical virology generating numerous novel assays with an obvious impact on the care of immunosuppressed and transplant patients. Cell culture-based assays for C. trachomatis were fully replaced by amplification and for all clinically relevant microorganisms multiple sensitive and specific PCR tests were developed, some of which are now fully automated [6, 30]. Real point-of-care tests for difficult-to-detect organisms including Mycobacterium tuberculosis, are currently available, generating a result within the hour without any complicated sample processing steps [31]. Even the difficult-to-diagnose syndromes such as bacterial sepsis and pneumonia can be addressed using some form or another of molecular testing [32, 33]. In addition, specific tests for a variety of virulence or anti-microbial resistance genes have been developed in large numbers. Several of these tests have been cleared by the American Food and Drug Administration (FDA) and are hence ready to fulfill their central role in laboratory innovation [34, 35]. Clearly, PCR is here to stay, although broad acceptance of many of the PCR tests has not yet been fully realized. This also includes tests that monitor host polymorphisms that can predict risk of colonization and/or infection [36]. Such assays will undoubtedly advance and expand in number and quality over the coming decade. Technological improvements are continuous: the use of droplet PCR, for instance, allows for very accurate quantification of the number of target molecules in the clinical starting material [37, 38]. The availability of so-called aptamers, single stranded short RNA or DNA molecules that have exquisite affinity for only a single ligand may again significantly impact nucleic acid diagnostics [39]. We have certainly not yet witnessed the end of the impact that molecular tests will have in clinical bacteriology.
MS was recently introduced as a new diagnostic tool in clinical microbiology with a first successful application for the detection and sequence-based identification of PCR products [40, 41]. Although the biophysical technology is intrinsically complicated (and beyond the scope of this review), it has to be said that from a practical, laboratory-utilization perspective MALDI-TOF MS is quite simple from a performance standpoint. A bacterial colony grown on a solid agar medium needs to be carefully sampled with a calibrated plastic loop or a toothpick. The biomaterial is deposited on a sample holder and a matrix solution needs to be added. Simple and direct pre-spotting lysis protocols for yeasts, molds and mycobacteria are also available. The sample slide is positioned in the MS machine and bacterial species identification is accomplished by comparing the generated spectrum of proteins (and peptides) to defined mass spectra contained within a reference spectral database representing multiple strains of known bacterial species. The current databases contain hundreds to thousands of such reference spectra and species identification (ID) is reliable and fast. This is precisely why this technology was so quickly accepted by clinical microbiologists. After the introduction of serological and molecular methods, biophysics has now definitely and irreversibly entered the clinical microbiology laboratory.
MALDI-TOF MS is extremely well suited for species identification since the major mass profile that is generated comes primarily from ribosomal proteins, which aligns nicely with current taxonomic classification. However, other applications are being realized including epidemiological typing which is relatively easy to perform relative to other standard methods such as PFGE and multi-locus sequence typing (MLST) [42]. For the purpose of epidemiological typing of bacterial isolates, mass peaks that are specific for a given microbial strain are easily identified and can be used to develop binary typing systems. In addition, certain resistance mechanisms can be identified as well. In case of AST or antimicrobial resistance testing (ART), degradation products of beta-lactam antibiotics that were hydrolyzed by beta-lactamases have been identified in a number of independent studies [43-46]. By using different (mixtures of) beta-lactam antibiotics as substrates, adequate identification of beta-lactamases could be achieved. Cellular changes or modifications under the influence of antibiotics can be detected as well as was recently demonstrated for the interaction between Candida albicans cells versus fluconazole and Candida and Aspergillus species versus caspofungin [47, 48]. In addition, MS can also be used to identify PCR products derived from resistance genes [40]. The iPLEX MassArray assays developed by Sequenom (San Diego, CA, USA) provide a good example of this flexible technology. The use of reporter molecules for which the mass changes when in contact with microbial virulence factors has been described to help assess the putative invasive potential of bacterial species. Signal peptides that are specifically cut by known proteases have shown diagnostic value for the identification of anthrax and periodontitis [49, 50].
Of course there are numerous confounding issues associated with diagnostic MS. First, culture is required since the sensitivity of the systems is in the order of 104-105 cells per assay. These are numbers rarely encountered in clinical specimens, with the possible exception of patients with significant urinary tract infection [51]. Second, equipment is expensive, bulky, sometimes noisy and in need of costly maintenance. Third, the distinction between certain genera, species or pathovars (Escherichia coli and Shigella spp for example) is still hard to make [52]. This renders regular updates of the database a strict requirement.
DNA sequencing had already developed into a standard research tool before it entered into the clinical arena. The technology as developed by Sanger is still widely used to sequence relatively short stretches of (usually PCR-amplified) DNA. This has been particularly useful for 16S rDNA sequence identification of bacteria at the species level, but also for epidemiological purposes (MLST; for a review, see [53]). Sanger sequencing is frequently used to detect point mutations associated with resistance to antiviral compounds or antibiotics. However, the importance of sequencing became even more apparent with the advent of newer generation sequencing procedures in infectious disease diagnosis [54]. These methods for the first time allowed for full sequencing of entire bacterial genomes [55]. When whole genome sequencing is applied in the context of microbial epidemiology or antimicrobial resistance, it can help in generating refined dissemination maps as well as "resistomes" and "toxomes" [56, 57].
To date, the technology has advanced to a state where even the genomes of complex mixtures of bacteria can be determined [58, 59]. This so-called microbial metagenomics can be used to generate complete catalogues of genomic components of bacterial species present in a variety of clinical materials including fecal specimens. In turn, this enables the R&D laboratory to detect differences in the composition of the bacterial flora in healthy versus diseased individuals. This will surely identify species that may be causal in syndromes as different as asthma, Crohn's disease and cardiovascular afflictions [60], although to date it is not clear when this technology will become routine in the clinical microbiology laboratory. Metagenomics is essential in trying to understand the overall effect of antibiotic treatment on the human microbiome as a whole [61]. At a more advanced state, next generation sequencing (NGS) will also facilitate genetic testing of host susceptibility towards infection or colonization. These current basic research developments will become more visible in the diagnostic laboratory over the coming decade.
The commercial availability of DNA sequencing instrumentation is already large. Companies such as Illumina, Roche and Ion Torrent offer affordable laboratory equipment with a limited footprint and simplified physics and chemistry [62]. The fact that some of these technologies in the end will also enable sequencing of additional biopolymers (proteins, oligo-and polysaccharides and fatty acids) and probably even detection and identification of small molecules of a diverse chemical nature will result in even more accelerated market penetration of unprecedented diagnostic assays.
In addition to the more classical diagnostic microbiology approaches there are a number of novel technologies that may be of significance in the not-too-far-away future. Some of these are highlighted below and their potential diagnostic relevance is discussed.
The diagnostic relevance of volatile organic compounds (VOCs) has been described often in literature [63]. The problem with this approach has always been the limited reproducibility of the diagnostic assays and the difficulties encountered in concentration and/or efficient capture of the volatile compounds to be detected and/or identified. In addition, the equipment required was and generally still is complicated to work with, bulky and expensive. A cheap alternative (depending on eating habits) may depend on animals that can differentiate specific scents [64] although that is still considered controversial: having bulky rats with 50 cm tails perform clinical investigations will change the perception of "clinical rounds" in general. Hence, many diagnostic tests have been described but essentially none of them have attained routine status. However, the promise of direct breath testing of infected individuals could open innovative diagnostic avenues in the future.
The most widely used VOC detection procedures encompass micro-weighing techniques using vibrational methods, gas chromatography and changes in electrical conductivity of metal chips. The latter technology has recently been improved significantly with the design of systems that allowed for the kinetic measurement of VOC production by growing bacteria in the head space of an enclosed culture container. Using such approaches it has been demonstrated that the distinction of several bacterial species could be reliably made on the basis of their longitudinal VOC production profile [65]. Next to this dynamic and culture medium-dependent method, it also appears that direct assessment of disease specific tuberculosis markers is feasible. A field study in Bangladesh, among patients and closely matched controls revealed that patients could be identified with reasonable sensitivity and specificity (manuscript submitted). It was anticipated that a diagnostic tool could be produced for less than 100 euro, whereas the tool could be regenerated and used for the diagnosis of multiple patients.
This would suggest that the detection of VOCs could be developed into point of care assays that would possibly enable improved treatment procedures for those patients in geographic regions where this kind of care is most needed. It is interesting to note that for M. tuberculosis a recent paper touted the cataloguing of many if not all of the VOCs produced during in vitro growth of the organism. The combined uses of ion flow tube MS and thermal desorption gas chromatography MS helped identify 2-phenylethanol as a unique volatile marker for mycobacterial growth [66]. This work could potentially be translated into specific test for this or other organism-specific volatiles. In addition, these formats do show promise for antibiotic susceptibility testing in bacteriology [67].
There are two examples of vibrational/absorption-based technologies that have been addressed with equal frequency in recent literature. Infra-red spectroscopy depends on irradiation of biological specimens with infra-red light followed by measurement of characteristic absorption and transmission profiles [68]. The disadvantage of this technology is that water strongly contributes to the absorption spectra, which complicates the analysis. This may be why Raman spectroscopy has been favored slightly. Raman spectroscopy measures light scattering that takes place upon illumination of biological specimens with visible (laser) light [69]. The disadvantage in this case is that autofluorescent bacterial components can strongly interfere with spectrum generation (e.g. carotene in Staphylococcus aureus). A relatively pure preparation of bacteria containing a significant number of cells is required for either technology, although single cell applications have been described [70, 71]. We will only mention some examples of Raman applications, but we emphasize that infrared (IR) spectroscopy may be equally applicable if the water signal is efficiently suppressed.
The most "visible" contribution of Raman spectroscopy in clinical microbiology has been in the field of microbial epidemiology. Although it has repeatedly been claimed that Raman spectroscopy can be used to distinguish virtually all clinically relevant bacterial species, definitive data are thus far missing. However, several studies concluded convincingly that typing of different strains belonging to a single species can be performed adequately. For various species of Staphylococcus, Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa, it has been demonstrated that Raman-mediated typing generates results that are concordant with those of current gold standards [72-75]. This has shown usefulness for delineating nosocomial outbreaks and even larger scale global dissemination of certain multi-drug resistant strains further suggesting that Raman (and IR) spectroscopy could be developed into useful diagnostic tools with additional research.
In conclusion, a variety of (biophysical) methods have been introduced into the clinical microbiology arena over the past decade. These methods clearly meet certain diagnostic requirements and development is surely continuing. In addition, the use of microfluidic tools will lead to miniaturization of equipment and more facile single cell handling. Such systems, when simplified to paper and lateral flow formats with targeted capillary capabilities, may be well suited for (low budget) field studies even in developing countries [76-79]. An excellent review on the emergence of such diagnostic nanotechnologies was published recently [80]. Some of these methods have been successfully introduced in the clinical microbiology laboratory which remains a matter of time for numerous other applications. For other methods, e.g., impedance sensing, there is only a proof of principle study at present with no information regarding feasibility [81]. Some studies, however, have demonstrated useful results for both gram positive and negative bacterial identification [82, 83]. Evolution of detection and identification assays in clinical microbiology progresses swiftly.
The development and spread of multi-drug resistant microorganisms including pan- resistant strains generates a universal threat to both humans and animals [84]. Prevention of the development of pan-resistance seems impossible [85] given the environmental antibiotic-mediated selective pressure [86-88]. Because of this, the diagnostic microbiology field should be strategically aligned towards surveillance and early detection of such resistance. However, AST is lagging behind in continuous improvement as compared to microbial detection and identification. Where most microbiologists will agree that nucleic acidbased diagnostics and MS will dominate the detection/identification fields over the coming years, the prospects for AST improvements are relatively ill-defined. To date, AST is primarily manual, using classical tests such as the use of antibiotic-containing screening media, disk diffusion methodology or EtestR. While automation is available (VITEK2™, Phoenix, Microscan WalkAway etc.), the technology remains growth-dependent. Of course, with the availability of PCR, several systems for molecular AST have been proposed. Still, the "real time PCR antibiogram" is a research rather than a diagnostic tool [89]. A recent paper on the development of a refined molecular test concluded that inhibition of growth as determined by PCR could be successfully used for defining minimum inhibitory concentrations (MICs) of antibiotics [90]. Again, this still remains stuck in the research phase. For future generation methodologies, the experts seem to be agreeing that NGS and also "next generation" MS may help to develop this analytical field, but the time scales for implementation are not clear. It is for these reasons that "intermediate technologies", i.e., ones that can be implemented sooner and provide real progress with regard to the classical culture-based methods are eagerly awaited. Below and in random order, are a limited number of technologies that may in the end help solve the current status quo.
AST is a core component of clinical microbiology since it guides proper antimicrobial chemotherapy. Obviously, many simple tools for facilitating its interpretation have been developed. One of them is simple photography of plates where disk diffusion tests were applied. Camera systems have been commercialized and these show useful efficacy [91]. This was extended for use with micro-colonies as well. Serial photography generated mini-movies of (non-)growing micro-colonies in the selective presence or absence of various antibiotics. The BACcel™ system as manufactured by Accelr8 is a good example of a machine that can do a one day AST for bacteria grown in blood culture bottles [92]. It shows that simple test formats can still be adapted into systems that serve the clinical microbiologist's needs in a better way.
Fluorescence activated cell sorting or FACS enables distinction of cells with different sizes and distinct levels of fluorescence. The fluorescence can be intrinsic to the cells or it can be specifically attached to or introduced in subsets of cells. For the first application, fluorescent antibodies are frequently used [93]. There are also fluorescent stains that penetrate only dead cells or cells with a permeable membrane [94]. Hence, cells that are affected or killed by antibiotics can potentially be distinguished from viable ones. During susceptibility testing, the ratio between dead and/or non-dividing cells changes over time and the use of fluorescent labeling and FACS can then help to determine the bactericidal or bacteriostatic effect of the antibiotic studied. It has been shown that methicillin susceptible S. aureus (MSSA) and methicillin resistant S. aureus (MRSA) can be distinguished by FACS after two-hour incubation with oxacillin [95]. When using micro-fluidic methods in combination with FACS, one can also distinguish cellular shapes and sizes. Although this has only been explored for red blood cells, the authors were convinced that this FACS approach could be used to distinguishing resistant from susceptible bacteria in the absence or presence of certain antibiotics [96]. Obviously, this technology is still far away from direct clinical application.
Cantilevers containing small canals which facilitate bacterial passage can be made to vibrate continuously. When bacteria pass such a system their sheer weight (in the femtogram range) will cause a change in the frequency of cantilever movement [97]. The nature of the change is correlated with the weight of the passing cells and "light" cells will cause a change in vibration that differs from that induced by "heavy" cells. When cells are treated with certain antiseptics or antibiotics or when they are subjected to unfavorable environmental conditions such as osmotic shock, their buoyant mass density may change and these changes can be measured [98]. This has been demonstrated for ampicillin resistant and susceptible variants of Citrobacter rodentium. It was also shown that resuscitation after osmotic shock for both variants and in the presence or absence of ampicillin could be easily differentiated in a short time-span. The method could potentially allow determination of the growth rate of bacterial cells having a doubling time of ten hours in a mere ten minutes. Cantilevers can be multiplexed using nanotechnology such that multiple antibiotics in various concentrations could be tested for a single growing culture simultaneously.
Bacteria produce energy and the level of production is affected by the presence of antibiotics to which the microbes may be susceptible. In case of resistance, steady-state energy production will be different in the presence of antibiotics. Recent studies in tuberculosis have shown the usefulness of isothermal micro-calorimetry for the rapid detection of mycobacteria and their susceptibility to isoniazid, ethambutol, and moxifloxacin [99]. The maximum bacterial growth rate and the lag phase were quantified by integrated heat flow-versus-time analysis. The micro-calorimetric technology used consisted of broadly affordable and sensitive micro-calorimeters. Also using isothermal micro-calorimetry, bacterial species identification from urine specimens was suggested to be possible even at low bacterial counts within a little over three hours on the basis of dynamic heat flow patterns [100].
Chip calorimetry is a monitoring tool for determining the physiological state of biofilms. Its potential use for the study of the effects of antibiotics was tested using an established model. The real-time monitoring potential of chip calorimetry was successfully demonstrated: a dosage of antibiotics initially increased the heat production rate probably due to activity of energy-dependent resistance mechanisms [101]. The subsequent reduction in heat production was probably due to the loss of activity and the death of the biofilm bacteria. This new analytical tool provided fast, quantitative, and mechanistic insights into the effects of antibiotics on biofilm activity. Detailed studies on the usefulness of this technology on AST in general are ongoing in various institutions worldwide.
Using optical tweezers capturing little gold nanoparticles and a microscopic sound source, it was recently shown that differences in the vibrational and, hence, energy level of the gold particles could be efficiently measured in liquid media [102]. The gold particle served as a specific "nano-ear" in this model system. Replacing the gold particle by a bacterial cell, either resistant or susceptible to a given antibiotic, in liquid media with or without antibiotics would facilitate characterization of the energy state of the single cells which is supposed to vary on the basis of phenotype and the presence or absence of antibiotics. The latter has not yet been convincingly demonstrated but further developments are eagerly awaited since this would provide another method for measuring the reactivity of single bacterial cells towards different antibiotics.
When magnetic beads are brought into specific magnetic fields they adapt a specific rotational spin. The frequency of rotation can be influenced by the binding of other molecules, viruses or bacteria. So if the beads are equipped with a ligand that specifically captures bacterial cells, the rotation of the beads changes at the moment of capture. This change can be measured. If one would pair all beads in a broth culture with one or two cells, which can be done by incubating derivatized beads with a diluted bacterial suspension and then wash, they will assume a constant rotational frequency. If the bacteria start to divide, the rotation frequency starts to change. If this division can be inhibited or blocked by antibiotics (that is, if the cells are susceptible to the antibiotic applied) then the change will not take place. If the bacteria are resistant to the (amount of) antibiotic applied, then again the change in rotational frequency will take place. In this way bacterial resistance can be determined and precisely quantified. Interestingly, specific formats of magnetic nanosensors can also be used to diagnose the presence of intracellular pathogens in host cell populations [103]. Novel developments in this field comprise the use of self-assembling magnetic particles that further facilitate efficient MIC measurements [104].
In the recent past it has been demonstrated that pairs of bacterial strains that differ in AST profiles can be distinguished on the basis of the transcriptome generated in the presence or absence of antibiotics [105]. Antibiotic exposure induces a stereotypical change in transcriptional response within a few minutes [106] and genes involved in for instance SOS response were significantly up- or down-regulated [15, 107]. Even more recent data show that similar transcriptional changes can also be monitored for bacteria present in clinical materials [15]. Again, antibiotic pulsing revealed specific transcriptional signatures for resistant versus susceptible bacteria and mecA or vanA transcription could be detected for MRSA and vancomycin resistant enterococci (VRE), respectively. Still, these investigations are in early developmental stages and further validation needs to be performed. The downside of this approach is that the presence of heteroresistant subpopulations might not be detected amongst the SOS signals generated by the susceptible population.
Little milli-, micro- or nano-droplets can be used as small, individual reactors. When the droplets can be individually manipulated and when they contain bacteria in sufficient numbers of cells, these microenvironments can serve as mini-fermenters where metabolic activity and viability of cells can be monitored. Development of such a system became feasible once the emulsification process was controlled and the long term kinetic stability of the droplets could be guaranteed [108]. The system consisted of 100 nL droplets containing 103 bacteria per droplet and in different droplets, different concentrations of antibiotics could be established [109]. By following the droplets over time by epi-fluorescence measurement, survival or death could be monitored for the bacteria populating the different droplets. Even more recently droplets were prepared that contain single bacteria [110]. This technology can be miniaturized, easily multiplexed with respect to the number of antibiotics tested per bacterial strain and duration of the test can be as short as a single or a few bacterial replication cycles. Obviously, assessing technical reproducibility and the development of adequate reference MIC databases will take many years.
Bacteriophages need living bacteria for their replication such that bacteria that are susceptible to certain antibiotics cannot support the replication of bacteriophage in the presence of these antibiotics. In 1997, the first bacteriophage-based AST system was presented for Mycobacterium tuberculosis [111]. Two years later a clinical validation study of six different antibiotics was published [112] and it was demonstrated that a 90% drop in phage counts could be used as an indicator for the MIC. The system was optimized by including recombinant phages containing luciferase genes. Instead of doing phage counts, simple luminescence assays were developed [113]. It was shown that the luciferase assay could be done at low costs in two days as opposed to the average of 10 days for classical AST for M. tuberculosis. These data were independently replicated and technological innovation continued [114-116]. Some of the tests were even applied successfully in developing countries [117]. The system was even further refined by using "fluoromycobacteriophages", phages with intrinsic fluorescence [118] or by the application of real time PCR detection of (non-)replicating bacteriophages [119]. Interestingly, the system was never widely accepted in clinical microbiology, nor was it expanded to other bacterial applications. Two of the major problems were laboratory contamination with bacteriophage [120] and possible phage resistance of certain bacterial strains. So irrespective of the sound biological principle of this approach, this system is unlikely to change our AST practice.
In conclusion, classical microbiological methods for AST can still be successfully applied but then become the time limiting step in the clinical microbiology laboratory. Innovation in the field of AST in particular supported by microfluidic approaches [121-123] as described above, is urgently needed to accelerate clinical testing and improve medical decision making with regard to antimicrobial therapy. Obviously, genome sequencing will be considered for novel generations of AST formats and the first examples linking genome sequencing with the identification of appropriate antimicrobial therapy has already been published [124]. As another example, programmed cell death in bacteria subjected to antimicrobials, highly sophisticated methods including nuclear magnetic resonance (NMR) or assessment of dielectric permittivity may help identify novel metabolic or biophysical markers of susceptibility or resistance [125-127]. However, as for all of the other innovative methods, setting up optimal reference databases will take significant R&D efforts for the years to come.
Full or partial microbiology laboratory automation [128, 129], depending on the size of the laboratory, is important to increase the speed of species identification and to accelerate AST. This philosophy can be strongly supported by dedicated use of smartphones or tablet computers [130]. Nucleic acid-mediated diagnostic methods are with us to stay and are slowly getting their much deserved recognition. At the same time, MS is revolutionizing post-culture bacterial identification. Although we are far from direct processing of clinical specimens using this technology, much research effort will be focused toward that end in the coming years. Additional biophysical methodologies, accompanied by the widely acknowledged "omics" technologies (genomics, transcriptomics, metabolomics, glycomics, lipidomics, interactomics etc.), are slowly being introduced and there is a healthy research focus on single cell detection and manipulation [131]. Human markers for colonization and infection susceptibility will steadily be added to the diagnostic repertoire over the coming years (next to the currently available C-reactive protein [132], procalcitonin [133] and others [134]) and laboratory automation will continue to evolve [135]. From that perspective, the future microbiologist should acquire significant knowledge in the fields of (large scale) data management, bioinformatics and communication. Challenging times lay ahead of us.
References
1. O'hara CM. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev. 2005; 18:147–162. PMID: 15653824.
2. Treviño M, Areses P, Peñalver MD, Cortizo S, Pardo F, del Molino ML, et al. Susceptibility trends of Bacteroides fragilis group and characterisation of carbapenemase-producing strains by automated REP-PCR and MALDI-TOF. Anaerobe. 2012; 18:37–43. PMID: 22261518.
3. Saravolatz LD, Pawlak J, Johnson LB. In Vitro susceptibilities and molecular analysis of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus isolates. Clin Infect Dis. 2012; 55:582–586. PMID: 22615331.
4. Steingart KR, Henry M, Laal S, Hopewell PC, Ramsay A, Menzies D, et al. Commercial serological antibody detection tests for the diagnosis of pulmonary tuberculosis: a systematic review. PLoS Med. 2007; 4:e202. PMID: 17564490.

5. Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol. 2011; 77:7877–7885. PMID: 21856826.
6. Fredlund H, Falk L, Jurstrand M, Unemo M. Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS. 2004; 112:771–784. PMID: 15638837.
7. Hadgu A, Dendukuri N, Hilden J. Evaluation of nucleic acid amplification tests in the absence of a perfect gold-standard test: a review of the statistical and epidemiologic issues. Epidemiology. 2005; 16:604–612. PMID: 16135935.
8. Van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today's and tomorrow's clinical microbiology laboratories. J Clin Microbiol. 2012; 50:1513–1517. PMID: 22357505.

9. Zhang W, Li F, Nie L. Integrating multiple 'omics' analysis for microbial biology: application and methodologies. Microbiology. 2010; 156:287–301. PMID: 19910409.

10. Kerremans JJ, Verbrugh HA, Vos MC. Frequency of microbiologically correct antibiotic therapy increased by infectious disease consultations and microbiological results. J Clin Microbiol. 2012; 50:2066–2068. PMID: 22422850.

11. Galar A, Keiva J, Espinosa M, Guillen-Grima F, Hernaez S, Yuste JR. Clinical and economic evaluation of the impact of rapid microbiological diagnostic testing. J Infect. 2012; 6. 19. [Epub ahead of print].

12. Kerremans JJ, van der Bij AK, Goessens W, Verbrugh HA, Vos MC. Needle-to-incubator transport time: logistic factors influencing transport time for blood culture specimens. J Clin Microbiol. 2009; 47:819–822. PMID: 19129412.

13. Griffiths LJ, Anyim M, Doffman SR, Wilks M, Millar MR, Agrawal SG. Comparison of DNA extraction methods for Aspergillus fumigatus using real-time PCR. J Med Microbiol. 2006; 55:1187–1191. PMID: 16914647.
14. Dauphin LA, Walker RE, Petersen JM, Bowen MD. Comparative evaluation of automated and manual commercial DNA extraction methods for detection of Francisella tularensis DNA from suspensions and spiked swabs by real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2011; 70:299–306. PMID: 21546201.
15. Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A. 2012; 109:6217–6222. PMID: 22474362.

16. Ingham C, Schneeberger . Hays , Van Leeuwen , editors. Can we improve on the Petri dish with porous culture support? Bentham Science Publisher eBook series. 2012. p. 3–16.
17. Ingham CJ, Boonstra S, Levels S, de Lange M, Meis JF, Schneeberger PM. Rapid susceptibility testing and microcolony analysis of Candida spp. cultured and imaged on porous aluminum oxide. PLoS One. 2012; 7:e33818. PMID: 22439000.
18. Váradi L, Gray M, Groundwater PW, Hall AJ, James AL, Orenga S, et al. Synthesis and evaluation of fluorogenic 2-amino-1,8-naphthyridine derivatives for the detection of bacteria. Org Biomol Chem. 2012; 10:2578–2589. PMID: 22354016.

19. Van Hoecke F, Deloof N, Claeys G. Performance evaluation of a modified chromogenic medium, ChromID MRSA New, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. Eur J Clin Microbiol Infect Dis. 2011; 30:1595–1598. PMID: 21499707.
20. Nordmann P, Girlich D, Poirel L. Detection of carbapenemase producers in enterobacteriaceae by use of a novel screening medium. J Clin Microbiol. 2012; 50:2761–2766. PMID: 22357501.

21. Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum beta-lactamase production by Enterobacteriaceae. J Clin Microbiol. 2011; 49:1048–1057. PMID: 21248086.
22. London R, Schwedock J, Sage A, Valley H, Meadows J, Waddington M, et al. An automated system for rapid non-destructive enumeration of growing microbes. PLoS One. 2010; 5:e8609. PMID: 20062794.

23. Towns ML, Jarvis WR, Hsueh PR. Guidelines on blood cultures. J Microbiol Immunol Infect. 2010; 43:347–349. PMID: 20688297.

24. Jin WY, Jang SJ, Lee MJ, Park G, Kim MJ, Kook JK, et al. Evaluation of VITEK2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diagn Microbiol Infect Dis. 2011; 70:442–447. PMID: 21767700.
25. Doern GV, Scott DR, Rashad AL. Clinical impact of rapid antimicrobial susceptibility testing of blood culture isolates. Antimicrob Agents Chemother. 1982; 21:1023–1024. PMID: 7114834.

26. Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008; 61:428–435. PMID: 18156278.

27. Horn JE, Quinn T, Hammer M, Palmer L, Falkow S. Use of nucleic acid probes for the detection of sexually transmitted infectious agents. Diagn Microbiol Infect Dis. 1986; 4(3 Suppl):101S–109S. PMID: 3084160.

28. Mulks MH, Simpson DA, Shoberg RJ. Restriction site polymorphism in genes encoding type 2 but not type 1 gonococcal IgA1 proteases. Antonie Van Leeuwenhoek. 1987; 53:471–478. PMID: 2897189.

29. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003; 41:1574–1585. PMID: 12682148.
30. Rank EL, Sautter RL, Beavis KG, Harris S, Jones S, Drechsel R, et al. A two-site analytical evaluation of the BD Viper System with XTR technology in non-extracted mode and extracted mode with seeded simulated specimens. J Lab Autom. 2011; 16:271–275. PMID: 21764022.

31. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010; 363:1005–1015. PMID: 20825313.

32. Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, et al. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn. 2010; 10:399–415. PMID: 20465496.

33. Tenover FC. Developing molecular amplification methods for rapid diagnosis of respiratory tract infections caused by bacterial pathogens. Clin Infect Dis. 2011; 52(Suppl 4):S338–S345. PMID: 21460293.

34. Emmadi R, Boonyaratanakornkit JB, Selvarangan R, Shyamala V, Zimmer BL, Williams L, et al. Molecular methods and platforms for infectious diseases testing a review of FDA-approved and cleared assays. J Mol Diagn. 2011; 13:583–604. PMID: 21871973.
35. van Hal SJ, Jennings Z, Stark D, Marriott D, Harkness J. MRSA detection: comparison of two molecular methods (BD GeneOhm PCR assay and Easy-Plex) with two selective MRSA agars (MRSA-ID and Oxoid MRSA) for nasal swabs. Eur J Clin Microbiol Infect Dis. 2009; 28:47–53. PMID: 18663499.

36. Van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009; 9:32–47. PMID: 19000784.
37. Angione SL, Chauhan A, Tripathi A. Real-time droplet DNA amplification with a new tablet platform. Anal Chem. 2012; 84:2654–2661. PMID: 22320164.

38. Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, et al. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip. 2011; 11:3838–3845. PMID: 21959960.

39. DeGrasse JA. A single-stranded DNA aptamer that selectively binds to Staphylococcus aureus enterotoxin B. PLoS One. 2012; 7:e33410. PMID: 22438927.
40. Syrmis MW, Moser RJ, Whiley DM, Vaska V, Coombs GW, Nissen MD, et al. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2011; 17:1804–1810. PMID: 21595795.
41. Ivanov PL. A new approach to forensic medical typing of human mitochondrial DNA with the use of mass-spectrometric analysis of amplified fragments: PLEX-ID automated genetic analysis system. Sud Med Ekspert. 2010; 53:46–51. PMID: 20734792.
42. Shitikov E, Ilina E, Chernousova L, Borovskaya A, Rukin I, Afanas'ev M, et al. Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect Genet Evol. 2012; 12:838–845. PMID: 22230718.

43. Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011; 49:3222–3227. PMID: 21775535.

44. Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol. 2011; 49:3321–3324. PMID: 21795515.

45. Hooff GP, van Kampen JJ, Meesters RJ, van Belkum A, Goessens WH, Luider TM. Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J Proteome Res. 2012; 11:79–84. PMID: 22013912.

46. Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 Carbapenemases by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol. 2012; 50:2441–2443. PMID: 22553235.
47. Marinach C, Alanio A, Palous M, Kwasek S, Fekkar A, Brossas JY, et al. MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics. 2009; 9:4627–4627. PMID: 19750514.
48. De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol. 2012; 50:2479–2483. PMID: 22535984.
49. Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman EC, et al. Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol. 2012; 50:104–112. PMID: 22075590.
50. Kaman WE, Hulst AG, van Alphen PT, Roffel S, van der, Merkel T, et al. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal Chem. 2011; 83:2511–2517. PMID: 21370823.
51. Köhling HL, Bittner A, Müller KD, Buer J, Becker M, Rübben H, et al. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J Med Microbiol. 2012; 61:339–344. PMID: 22275503.

52. Everley RA, Mott TM, Wyatt SA, Toney DM, Croley TR. Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J Am Soc Mass Spectrom. 2008; 19:1621–1628. PMID: 18692404.
53. Turner KM. The secret life of the multilocus sequence type. Int J Antimicrob Agents. 2007; 29:129–135. PMID: 17204401.

54. Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011; 365:347–357. PMID: 21793746.

55. Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995; 269:496–512. PMID: 7542800.
56. Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012; 2:e001124.
57. Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012; 366:2267–2275. PMID: 22693998.

58. Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O'Huallachain M, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2011; 30:78–82. PMID: 22178993.

59. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2011; 13:47–58. PMID: 22179717.

60. Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008; 24:4–10. PMID: 18043225.

61. Sommer MO, Dantas G. Antibiotics and the resistant microbiome. Curr Opin Microbiol. 2011; 14:556–563. PMID: 21802347.

62. Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012; 30:434–439. PMID: 22522955.

63. Korpi A, Järnberg J, Pasanen AL. Microbial volatile organic compounds. Crit Rev Toxicol. 2009; 39:139–193. PMID: 19204852.

64. Mgode GF, Weetjens BJ, Nawrath T, Cox C, Jubitana M, Machang'u RS, et al. Diagnosis of tuberculosis by trained African giant pouched rats and confounding impact of pathogens and microflora of the respiratory tract. J Clin Microbiol. 2012; 50:274–280. PMID: 22135255.

65. Bruins M, Bos A, Petit PL, Eadie K, Rog A, Bos R, et al. Device-independent, real-time identification of bacterial pathogens with a metal oxide-based olfactory sensor. Eur J Clin Microbiol Infect Dis. 2009; 28:775–780. PMID: 19190942.

66. McNerney R, Mallard K, Okolo PI, Turner C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol Lett. 2012; 328:150–156. PMID: 22224870.

67. Crespo E, Cristescu SM, de Ronde H, Kuijper S, Kolk AH, Anthony RM, et al. Proton Transfer Reaction Mass Spectrometry detects rapid changes in volatile metabolite emission by Mycobacterium smegmatis after the addition of specific antimicrobial agents. J Microbiol Methods. 2011; 86:8–15. PMID: 21277343.
68. Beekes M, Lasch P, Naumann D. Analytical applications of Fourier transform-infrared (FT-IR) spectroscopy in microbiology and prion research. Vet Microbiol. 2007; 123:305–319. PMID: 17540519.

69. Maquelin K, Kirchner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods. 2002; 51:255–271. PMID: 12223286.

70. Shachaf CM, Elchuri SV, Koh AL, Zhu J, Nguyen LN, Mitchell DJ, et al. A novel method for detection of phosphorylation in single cells by surface enhanced Raman scattering (SERS) using composite organic-inorganic nanoparticles (COINs). PLoS One. 2009; 4:e5206. PMID: 19367337.

71. Orsini F, Ami D, Villa AM, Sala G, Bellotti MG, Doglia SM. FT-IR microspectroscopy for microbiological studies. J Microbiol Methods. 2000; 42:17–27. PMID: 11000427.

72. Willemse-Erix D, Bakker-Schut T, Slagboom-Bax F, Jachtenberg JW, Lemmens-den Toom N, Papagiannitsis CC, et al. Rapid typing of extended-spectrum β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates by use of SpectraCell RA. J Clin Microbiol. 2012; 50:1370–1375. PMID: 22238437.
73. Wulf MW, Willemse-Erix D, Verduin CM, Puppels G, van Belkum A, Maquelin K. The use of Raman spectroscopy in the epidemiology of methicillin-resistant Staphylococcus aureus of human- and animal-related clonal lineages. Clin Microbiol Infect. 2012; 18:147–152. PMID: 21854500.
74. Willemse-Erix DF, Jachtenberg JW, Schut TB, van Leeuwen W, van Belkum A, Puppels G, et al. Towards Raman-based epidemiological typing of Pseudomonas aeruginosa. J Biophotonics. 2010; 3:506–511. PMID: 20572285.
75. Willemse-Erix HF, Jachtenberg J, Barutçi H, Puppels GJ, van Belkum A, Vos MC, et al. Proof of principle for successful characterization of methicillin-resistant coagulase-negative staphylococci isolated from skin by use of Raman spectroscopy and pulsed-field gel electrophoresis. J Clin Microbiol. 2010; 48:736–740. PMID: 20042618.

76. Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011; 17:1015–1019. PMID: 21804541.

77. Joon Tam Y, Mohd Lila MA, Bahaman AR. Development of solid - based paper strips for rapid diagnosis of Pseudorabies infection. Trop Biomed. 2004; 21:121–134. PMID: 16493404.
78. Thornton CR. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin Vaccine Immunol. 2008; 15:1095–1105. PMID: 18463222.

79. Lafleur L, Stevens D, McKenzie K, Ramachandran S, Spicar-Mihalic P, Singhal M, et al. Progress toward multiplexed sample-to-result detection in low resource settings using microfluidic immunoassay cards. Lab Chip. 2012; 12:1119–1127. PMID: 22311085.

80. Shinde SB, Fernandes CB, Patravale VB. Recent trends in in-vitro nanodiagnostics for detection of pathogens. J Control Release. 2011; 159:164–180. PMID: 22192572.

81. Siddiqui S, Dai Z, Stavis CJ, Zeng H, Moldovan N, Hamers RJ, et al. A quantitative study of detection mechanism of a label-free impedance biosensor using ultrananocrystalline diamond microelectrode array. Biosens Bioelectron. 2012; 35:284–290. PMID: 22456097.

82. Chang TC, Huang AH. Rapid differentiation of fermentative from non-fermentative gram-negative bacilli in positive blood cultures by an impedance method. J Clin Microbiol. 2000; 38:3589–3594. PMID: 11015369.

83. Wu JJ, Huang AH, Dai JH, Chang TC. Rapid detection of oxacillin-resistant Staphylococcus aureus in blood cultures by an impedance method. J Clin Microbiol. 1997; 35:1460–1464. PMID: 9163462.
84. Walsh TR, Toleman MA. The emergence of pan-resistant Gram-negative pathogens merits a rapid global political response. J Antimicrob Chemother. 2012; 67:1–3. PMID: 21994911.

85. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010; 74:417–433. PMID: 20805405.

86. Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2011; 8:6–9. PMID: 22173343.

87. Belenky P, Collins JJ. Microbiology. Antioxidant strategies to tolerate antibiotics. Science. 2011; 334:915–916. PMID: 22096180.
88. Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011; 473:216–220. PMID: 21562562.

89. Waldeisen JR, Wang T, Mitra D, Lee LP. A real-time PCR antibiogram for drug-resistant sepsis. PLoS One. 2011; 6:e28528. PMID: 22164303.

90. Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PF. Antibiotic susceptibility testing of grown blood cultures by combining culture and real-time polymerase chain reaction is rapid and effective. PLoS One. 2011; 6:e27689. PMID: 22194790.

91. Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis. 2009; 49:1749–1755. PMID: 19857164.

92. Douglas IS, Price C, Overdier K, Thompson K, Wolken B, Metzger S, et al. Rapid Microbiological Identification and Major Drug Resistance Phenotyping with Novel Multiplexed Automated Digital Microscopy (MADM) for Ventilator-associated Pneumonia (VAP) Surveillance. 2011. Updated in 2012. In : American Thoracic Society; Denver, CO.. http://www.accelr8.com/docs/ATS_2011a.pdf.
93. Cronin UP, Wilkinson MG. The potential of flow cytometry in the study of Bacillus cereus. J Appl Microbiol. 2010; 108:1–16. PMID: 19486207.
94. Filoche SK, Coleman MJ, Angker L, Sissons CH. A fluorescence assay to determine the viable biomass of microcosm dental plaque biofilms. J Microbiol Methods. 2007; 69:489–496. PMID: 17408789.

95. Shrestha NK, Scalera NM, Wilson DA, Procop GW. Rapid differentiation of methicillin-resistant and methicillin-susceptible Staphylococcus aureus by flow cytometry after brief antibiotic exposure. J Clin Microbiol. 2011; 49:2116–2120. PMID: 21471343.
96. Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Sorting cells by size, shape and deformability. Lab Chip. 2012; 12:1048–1051. PMID: 22327631.

97. Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, et al. Using buoyant mass to measure the growth of single cells. Nat Methods. 2010; 7:387–390. PMID: 20383132.

98. Knudsen SM, von Muhlen MG, Schauer DB, Manalis SR. Determination of bacterial antibiotic resistance based on osmotic shock response. Anal Chem. 2009; 81:7087–7090. PMID: 20337387.

99. Howell M, Wirz D, Daniels AU, Braissant O. Application of a micro-calorimetric method for determining drug susceptibility in Mycobacterium species. J Clin Microbiol. 2012; 50:16–20. PMID: 22090404.
100. Bonkat G, Braissant O, Widmer AF, Frei R, Rieken M, Wyler S, et al. Rapid detection of urinary tract pathogens using micro-calorimetry principle, technique and first results. BJU Int. 2012; 2. 07. [Epub ahead of print].

101. Buchholz F, Wolf A, Lerchner J, Mertens F, Harms H, Maskow T. Chip calorimetry for fast and reliable evaluation of bactericidal and bacteriostatic treatments of biofilms. Antimicrob Agents Chemother. 2010; 54:312–319. PMID: 19822705.

102. Ohlinger A, Deak A, Lutich AA, Feldmann J. Optically trapped gold nanoparticle enables listening at the microscale. Phys Rev Lett. 2012; 108:018101. PMID: 22304294.

103. Kaittanis C, Boukhriss H, Santra S, Naser SA, Perez JM. Rapid and sensitive detection of an intracellular pathogen in human peripheral leukocytes with hybridizing magnetic relaxation nanosensors. PLoS One. 2012; 7:e35326. PMID: 22496916.

104. Kinnunen P, McNaughton BH, Albertson T, Sinn I, Mofakham S, Elbez R, et al. Self-assembled magnetic bead biosensor for measuring bacterial growth and antimicrobial susceptibility testing. Small. 2012; 6. 05. [Epub ahead of print].

105. Fischer A, Yang SJ, Bayer AS, Vaezzadeh AR, Herzig S, Stenz L, et al. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J Antimicrob Chemother. 2011; 66:1696–1711. PMID: 21622973.
106. Sangurdekar DP, Srienc F, Khodursky AB. A classification based framework for quantitative description of large-scale microarray data. Genome Biol. 2006; 7:R32. PMID: 16626502.
107. Brazas MD, Hancock RE. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005; 49:3222–3227. PMID: 16048929.
108. Delmas T, Piraux H, Couffin AC, Texier I, Vinet F, Poulin P, et al. How to prepare and stabilize very small nanoemulsions. Langmuir. 2011; 27:1683–1692. PMID: 21226496.

109. Baraban L, Bertholle F, Salverda ML, Bremond N, Panizza P, Baudry J, et al. Millifluidic droplet analyser for microbiology. Lab Chip. 2011; 11:4057–4062. PMID: 22012599.

110. Boitard L, Cottinet D, Kleinschmitt C, Bremond N, Baudry J, Yvert G, et al. Monitoring single-cell bioenergetics via the coarsening of emulsion droplets. Proc Natl Acad Sci U S A. 2012; 109:7181–7186. PMID: 22538813.

111. Wilson SM, al-Suwaidi Z, McNerney R, Porter J, Drobniewski . Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat Med. 1997; 3:465–468. PMID: 9095184.
112. Eltringham IJ, Wilson SM, Drobniewski FA. Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999; 37:3528–3532. PMID: 10523547.
113. Banaiee N, Bobadilla-Del-Valle M, Bardarov S Jr, Riska PF, Small PM, Ponce-De-Leon A, et al. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J Clin Microbiol. 2001; 39:3883–3888. PMID: 11682502.
114. Hazbón MH, Guarín N, Ferro BE, Rodríguez AL, Labrada LA, Tovar R, et al. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003; 41:4865–4869. PMID: 14532245.
115. Banaiee N, Bobadilla-del-Valle M, Riska PF, Bardarov S Jr, Small PM, Ponce-de-Leon A, et al. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003; 52:557–561. PMID: 12808076.
116. Smietana M, Bock WJ, Mikulic P, Ng A, Chinnappan R, Zourob M. Detection of bacteria using bacteriophages as recognition elements immobilized on long-period fiber gratings. Opt Express. 2011; 19:7971–7978. PMID: 21643046.

117. Traore H, Ogwang S, Mallard K, Joloba ML, Mumbowa F, Narayan K, et al. Low-cost rapid detection of rifampicin resistant tuberculosis using bacteriophage in Kampala, Uganda. Ann Clin Microbiol Antimicrob. 2007; 6:1. PMID: 17212825.

118. Piuri M, Jacobs WR Jr, Hatfull GF. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS One. 2009; 4:e4870. PMID: 19300517.
119. Pholwat S, Ehdaie B, Foongladda S, Kelly K, Houpt E. Real-time PCR using mycobacteriophage DNA for rapid phenotypic drug susceptibility results for Mycobacterium tuberculosis. J Clin Microbiol. 2012; 50:754–761. PMID: 22170929.
120. Mole R, Trollip A, Abrahams C, Bosman M, Albert H. Improved contamination control for a rapid phage-based rifampicin resistance test for Mycobacterium tuberculosis. J Med Microbiol. 2007; 56:1334–1339. PMID: 17893170.
121. Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012; 46:561–572. PMID: 22633370.

122. Cira NJ, Ho JY, Dueck ME, Weibel DB. A self-loading microfluidic device for determining the minimum inhibitory concentration of antibiotics. Lab Chip. 2012; 12:1052–1059. PMID: 22193301.

123. Ding X, Lin SC, Kiraly B, Yue H, Li S, Chiang IK, et al. On-chip manipulation of single microparticles, cells, and organisms using surface acoustic waves. Proc Natl Acad Sci U S A. 2012; 109:11105–11109. PMID: 22733731.

124. Scholl D, Gebhart D, Williams SR, Bates A, Mandrell R. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One. 2012; 7:e33637. PMID: 22432037.

125. Contag CH. In vivo pathology: seeing with molecular specificity and cellular resolution in the living body. Annu Rev Pathol. 2007; 2:277–305. PMID: 18039101.

126. Rieder R, Zhao Z, Nittayajarn A, Zavizion B. Direct detection of the bacterial stress response in intact samples of platelets by differential impedance. Transfusion. 2011; 51:1037–1046. PMID: 20977486.

127. Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012; 7:166–171. PMID: 22007661.

128. Greub G, Prod'hom G. Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect. 2011; 17:655–660. PMID: 21521409.

129. Matthews S, Deutekom J. The future of diagnostic bacteriology. Clin Microbiol Infect. 2011; 17:651–654. PMID: 21521408.

130. Goff DA. iPhones, iPads and medical applications for antimicrobial stewardship. Pharmacotherapy. 2012; 32:657–661. PMID: 22605544.

131. Lin Y, Trouillon R, Safina G, Ewing AG. Chemical analysis of single cells. Anal Chem. 2011; 83:4369–4392. PMID: 21500835.

132. Mori Y, Miyawaki K, Kato K, Takenaka K, Iwasaki H, Harada N, et al. Diagnostic value of serum procalcitonin and C-reactive protein for infections after allogeneic hematopoietic stem cell transplantation versus non-transplant setting. Intern Med. 2011; 50:2149–2155. PMID: 21963733.

133. Fowler CL. Procalcitonin for triage of patients with respiratory tract symptoms: a case study in the trial design process for approval of a new diagnostic test for lower respiratory tract infections. Clin Infect Dis. 2011; 52(Suppl 4):S351–S356. PMID: 21460295.

134. Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. Immunological biomarkers of tuberculosis. Nat Rev Immunol. 2011; 11:343–354. PMID: 21475309.

135. Dumitrescu O, Dauwalder O, Lina G. Present and future automation in bacteriology. Clin Microbiol Infect. 2011; 17:649–650. PMID: 21521407.

Fig. 1
Schematic review of activities and procedures in the classical microbiology laboratory (green box). Essential pre-analytical and preparatory steps are given in light blue. Molecular technological innovation is indicated by the grey insertions in the green box, whereas the yellow box identifies host rather than infectious agent testing. The striped box in the middle relates to the recent but central position of novel technologies in the improvement of both host and agent specific testing which in the end should lead to personalization of infectious disease detection and treatment. Finally in dark blue: the central patient-oriented paradigm in the current evolution of microbial diagnostic services.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download