Abstract
Geosmithia argillacea, an anamorph of Talaromyces eburneus, is a thermophilic filamentous fungus that has a phenotype similar to that of the Penicillium species, except for the creamy-white colonies and cylindrical conidia. Recently, a new genus called Rasamsonia has been proposed, which is to accommodate the Talaromyces and Geosmithia species. Here, we report the first Korean case of G. argillacea isolated from a patient with a fungal ball. The patient was a 44-yr-old Korean man with a history of pulmonary tuberculosis and aspergilloma. The newly developed fungal ball in his lung was removed and cultured to identify the fungus. The fungal colonies were white and slow-growing, and the filaments resembled those of Penicillium. Molecular identification was carried out by sequencing the internal transcribed spacer (ITS) region of the 28S rDNA and the β-tubulin genes. A comparative sequence analysis using the GenBank (http://blast.ncbi.nlm.nih.gov/) database was performed with the basic local alignment search tool (BLAST) algorithm. The results revealed a 97-100% similarity with the G. argillacea ITS sequence. This case should increase awareness among physicians about the pathogenic potential of G. argillacea in humans and help them accurately identify this fungus, because it can be easily confused with Penicillium and Paecilomyces species owing to their similar phenotypic and microscopic characteristics. A molecular approach should be employed to enable accurate identification of G. argillacea.
Geosmithia argillacea, an anamorph of Talaromyces eburneus, is a thermophilic filamentous fungus that was first described in 1969 as a Penicillium sp. [1]. The genus Geosmithia was proposed by Pitt [2]. After comparison of the D1/D2 region of the 28S rDNA, Yaguchi et al. [3] concluded that Geosmithia eburnea (as Talaromyces eburneus) is a teleomorph of G. argillacea. Recently, a new genus Rasamsonia was established, which comprises both thermotolerant Talaromyces and thermophilic Geosmithia species [4].
Although its natural habitat remains unknown, airway colonization by G. argillacea has recently been reported in a few patients with cystic fibrosis who had previously received treatment with itraconazole with or without voriconazole [5, 6]. Intrinsically multidrug-resistant G. argillacea isolates may emerge as a cause of disseminated infections in chronic granulomatous disease patients receiving long-term azole therapy [7]. Moreover, a disseminated G. argillacea infection has also been reported in a patient with gastrointestinal graft-versus-host disease [8].
In Korea, there has been no previous report on G. argillacea infections. Here, we report a rare case of G. argillacea colonization as an intracavitary fungal ball in a patient with a history of pulmonary tuberculosis. The isolate was identified by comparative sequence analysis of its internal transcribed spacer (ITS) region and β-tubulin genes.
A 44-yr-old Korean man with recurrent pulmonary infection was admitted to the hospital for elective lung surgery. He was diagnosed with pulmonary tuberculosis at the age of 18; subsequently, he received antituberculous therapy and was cured. However, hemoptysis occurred 4 yr later due to the development of pulmonary aspergilloma, and a right upper lobectomy was performed. In July 2007, the hemoptysis recurred and nontuberculous Mycobacterium (NTM) species were detected in the sputum sample. The treatment for this NTM infection lasted for 2 yr. In November 2009, on high resolution computed tomography (HRCT) of the chest, and a newly formed foreign body, with features suggestive of an intracavitary fungal ball, was noted in his right lung. Mycobacterium tuberculosis was also detected in the sputum sample, and he received antituberculous medication for 9 months.
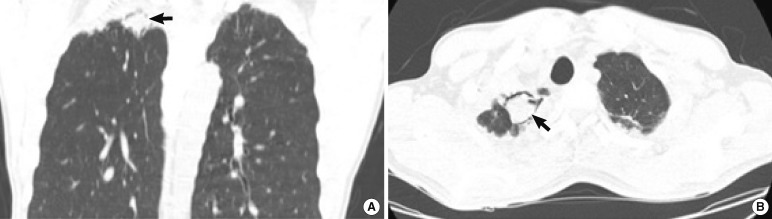
Several chest HRCT images taken between December 2010 and April 2011 revealed an increase in the size of the fungal ball, suggesting chronic necrotizing pulmonary aspergillosis (Fig. 1). Results of laboratory investigations were as follows: hemoglobin, 12.4 g/dL; white blood cell count, 6.03×109/L (50% segmented neutrophils, 32% lymphocytes, 16% monocytes, and 2% eosinophils). Serum electrolyte concentrations, liver function test results, and creatinine and glucose levels were all within the reference ranges. The serum sample was strongly positive for antibodies to Aspergillus, as determined by an enzyme immunoassay (>200 U/mL; positive cut-off: >12 U/mL). However, negative results were obtained for serum Aspergillus galactomannan. After the administration of antifungal therapy (itraconazole), the lesion was surgically resected in April 2011. Wide-wedge resection of the superior segment of the right lower lobe was performed along with the removal of the fungal ball. Simultaneously, lung tissue specimens were obtained for culture and pathology. Pathological study of the lung tissue revealed that the fungal ball was composed of hyphae, and therefore it was identified as an aspergilloma. Pathological evaluation revealed neither the presence of fungus in the lung parenchyma nor any evidence of invasive mycosis.
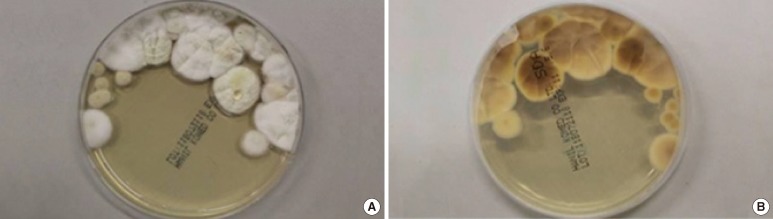
Lung tissue specimens were cultured on Sabouraud dextrose agar for 3 weeks, at a temperature of 30℃ for the first 2 days, and at room temperature (25℃) thereafter. Species was identified according to macro- and micro-morphological criteria. Macroscopic analysis revealed that the colonies formed were cream to beige in color, with no color on the reverse side (Fig. 2). Growth was slow and restricted at room temperature, but was enhanced at 30℃, with colonies reaching 2-3 cm in diameter after 10 days at this temperature. Microscopic analysis revealed the presence of hyaline, rough-walled, septate, and often-branched conidiophores (Fig. 3). These conidiophores bore biverticillate- or triverticillate-asymmetrical penicilli and had phialides with a tapering tip that were parallel to the axis. Conidia were smooth-walled, hyaline, and cylindrical to ellipsoidal. On the basis of these microscopic characteristics, it was assumed that the fungal ball was composed of a species of Penicillium. However, the macroscopic features were not compatible with those of Penicillium. Penicillium species are characterized by brush-shaped, round conidia and bluish-green colonies with a white border [9]. On the other hand, Paecilomyces was also excluded because the observed phialides were parallel to the axis of the conidiophores and were not bent away from the axis. Most importantly, Penicillium and Paecilomyces grow rapidly at room temperature.
Therefore, molecular identification was performed by sequencing the ITS region and β-tubulin genes. The ITS region and the β-tubulin gene fragments were amplified and sequenced by standard methods, according to the CLSI guidelines [10]. This was followed by sequence comparison with the GenBank (NCBI) (http://blast.ncbi.nlm.nih.gov/) database using the basic local alignment search tool (BLAST) algorithm. The BLAST search revealed that the partial ITS sequence showed a 100% (420/420) similarity with that of the G. argillacea strain CGDGA6 (accession no. HQ246728.1, #475-56), and a 97.1% (409/421) similarity with that of the G. argillacea type strain CBS 101.69 (accession no. JF417491.1, #497-79). Moreover, the β-tubulin gene sequence showed a 99.8% (423/424) similarity with that of the G. argillacea DTO 49D4 strain (accession no. GU968696.1, #1-424) and a 97.2% (424/436) similarity with that of the G. argillacea type strain CBS 101.69 (accession no. JF417491.1, #1-435). On the basis of these results, we concluded that G. argillacea was the species that was most likely responsible for the formation of fungal ball.
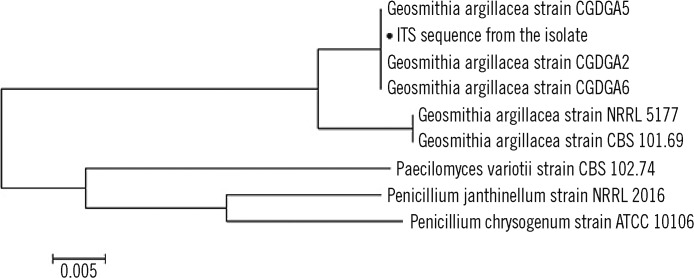
A phylogenetic tree was constructed using the neighbor-joining method on the basis of the results of the comparative sequence analysis of the ITS region (Fig. 4). Phylogenetic and molecular evolutionary analyses were conducted using the Molecular Evolutionary Genetics Analysis (MEGA) software version 5.0 (http://www.megasoftware.net) [11], and a subline related to Penicillium and Paecilomyces spp. was obtained, which are both similar to G. argillacea in microscopic findings.
Itraconazole was continued for another 2 months after surgery. Thereafter, the patient had a favorable clinical course without recurrence of symptoms or sequelae.
The first case of a G. argillacea infection was reported in 2009 in a German shepherd dog [12]. The pathogenic potential of G. argillacea in humans was first reported after it was detected in the sputum of patients with cystic fibrosis in 2010 [5, 6, 13]. G. argillacea is now considered one of the many respiratory pathogens associated with the pathophysiology of cystic fibrosis. It has been suggested that an immunocompromised state following lung transplantation might make a patient susceptible to a severe G. argillacea infection [5]. In 2011, various cases of invasive mycosis or disseminated diseases caused by G. argillacea were reported in patients with chronic granulomatous disease [7, 14] and gastrointestinal graft-versus-host disease [8]. These findings strongly suggest that infection with this fungus could be more severe in immunocompromised patients than in immunocompetent patients such as those with cystic fibrosis. In addition, multidrug resistant G. argillacea isolates may cause disseminated infections in patients with chronic granulomatous disease who receive long-term azole therapy [7].
The similarity in the phenotypic characteristics (e.g., thermophilicity) and microscopic morphology between G. argillacea and Penicillium species could be a barrier to the immediate and accurate identification of this fungus. According to our fungal culture protocol, clinical specimens should be incubated at 30℃ for the first 2 days and then at 25℃ on universal media (Sabouraud dextrose agar) for a maximum of 3 weeks. When incubated at room temperature (25℃), these creamy-white colonies showed a reduced growth rate and no conidia development upon microscopic observation. Even after 3 weeks, this mold could not be conclusively identified, but it was suspected to be a Penicillium species based on its microscopic morphology. When subcultured at 30℃ for 10 days, this strain showed typical macroscopic/microscopic morphologic characteristics of G. argillacea. Therefore, changing the culture conditions to those previously described would be helpful in accurate fungal identification, especially when the strain shows limited growth after using the standard protocol.
It has been suggested that the incidence of Geosmithia spp. isolation may be higher than that reported because this fungus may have been incorrectly identified as Penicillium or Paecilomyces [15]. This indicates that G. argillacea could be a more common human pathogen than previously estimated. Some studies support the presumption and have used direct sequencing methods to correctly implicate G. argillacea as the fungal pathogen in cases in which Paecilomyces variotii or unidentified isolates were previously identified as the causative pathogens [15, 16]. These reports also strongly suggest that molecular methods are very useful in fungal identification. Furthermore, antimicrobial susceptibility testing may be necessary to identify the appropriate treatment for a disseminated infection in immunocompromised patients [7]. However, there are still some limitations in the clinical implementation of antimicrobial susceptibility testing. Previous studies have shown that the results of in vitro antifungal susceptibility testing are not consistent with clinical responses [14]. Moreover, breakpoints are yet to be recommended by CLSI or other regulatory agencies. Supplementary studies to determine a relationship between clinical outcomes and antifungal susceptibility data are needed.
Interestingly, the serum Aspergillus antibody test was highly positive, but the serum Aspergillus galactomannan antigen test was negative. Before the fungus was identified, the findings from the antigen and antibody tests were considered to be indicative of allergic aspergillosis without invasive mycosis. After pulmonary aspergilloma had been ruled out, the positive Aspergillus antibody test was considered false positive due to immunological responses [17].
To the best of our knowledge, this is the first report of isolation of G. argillacea in Korea. This case should increase awareness among physicians about the pathogenic potential of G. argillacea in humans and will help them distinguish it from Penicillium and/or Paecilomyces species, which have similar phenotypic and microscopic characteristics. G. argillacea can colonize to form a fungal ball, manifesting as hemoptysis, especially in patients without underlying diseases (such as cystic fibrosis) that predispose them to respiratory pathogens and in immunocompromised patients (such as post-transplantation). Notably, the patient had an unusual history of recurrent pulmonary infections, including tuberculosis. An existing pulmonary cavity that was created during a previous lobectomy provided space for the fungal ball to grow. Although there was no evidence of immuno deficiency providing vulnerability to infection, we cannot exclude the possibility of the presence of a predisposing factor that promoted the colonization and/or infection by G. argillacea. Increased awareness among clinicians and microbiologists is necessary for them to fully comprehend the implications of a G. argillacea infection and understand the pathophysiology of this fungus.
References
1. Stolk AC, Evans HC, Nilsson T. Penicillium argillaceum sp.nov., a thermotolerant Penicillium. Trans Br Mycol Soc. 1969; 53:307–311.
2. Pitt JI. Geosmithia gen. nov. for Penicillium lavendulum and related species. Can J Bot. 1979; 57:2021–2030.
3. Yaguchi T, Udagawa S, Nishimura K. Geosmithia argillacea is the anamorph of Talaromyces eburneus as a heat resistant fungus. Cryptogam Mycol. 2005; 26:133–141.
4. Houbraken J, Spierenburg H, Frisvad JC. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek. 2012; 101:403–421. PMID: 21965082.
5. Giraud S, Pihet M, Razafimandimby B, Carrère J, Degand N, Mely L, et al. Geosmithia argillacea: an emerging pathogen in patients with cystic fibrosis. J Clin Microbiol. 2010; 48:2381–2386. PMID: 20463155.
6. Barton RC, Borman AM, Johnson EM, Houbraken J, Hobson RP, Denton M, et al. Isolation of the fungus Geosmithia argillacea in sputum of people with cystic fibrosis. J Clin Microbiol. 2010; 48:2615–2617. PMID: 20421435.
7. Machouart M, Garcia-Hermoso D, Rivier A, Hassouni N, Catherinot E, Salmon A, et al. Emergence of disseminated infections due to Geosmithia argillacea in patients with chronic granulomatous disease receiving long-term azole antifungal prophylaxis. J Clin Microbiol. 2011; 49:1681–1683. PMID: 21270214.
8. Valentin T, Neumeister P, Pichler M, Rohn A, Koidl C, Haas D, et al. Disseminated Geosmithia argillacea infection in a patient with gastrointestinal GvHD. Bone Marrow Transplant. 2012; 47:734–736. PMID: 21785470.
9. Larone DH, editor. Medically important fungi : a guide to identification. 2011. 5th ed. Washington, DC: ASM Press.
10. Clinical and Laboratory Standards Institute. Interpretive criteria for microorganism identification of bacteria and fungi by DNA target sequencing; approved guideline. Document MM18-A. 2008. Wayne PA: Clinical and Laboratory Standards Institute.
11. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739. PMID: 21546353.

12. Grant DC, Sutton DA, Sandberg CA, Tyler RD Jr, Thompson EH, Romanelli AM, et al. Disseminated Geosmithia argillacea infection in a German shepherd dog. Med Mycol. 2009; 47:221–226. PMID: 19169949.
13. Symoens F, Haase G, Pihet M, Carrere J, Beguin H, Degand N, et al. Unusual Aspergillus species in patients with cystic fibrosis. Med Mycol. 2010; 48:S10–S16. PMID: 21067321.
14. De Ravin SS, Challipalli M, Anderson V, Shea YR, Marciano B, Hilligoss D, et al. Geosmithia argillacea: an emerging cause of invasive mycosis in human chronic granulomatous disease. Clin Infect Dis. 2011; 52:e136–e143. PMID: 21367720.
15. Houbraken J, Verweij PE, Rijs AJ, Borman AM, Samson RA. Identification of Paecilomyces variotii in clinical samples and settings. J Clin Microbiol. 2010; 48:2754–2761. PMID: 20519470.
16. Jang JH, Lee JH, Ki CS, Lee NY. Identification of clinical mold isolates by sequence analysis of the internal transcribed spacer region, ribosomal large-subunit D1/D2, and β-tubulin. Ann Lab Med. 2012; 32:126–132. PMID: 22389879.

17. Kurup VP, Kumar A. Immunodiagnosis of aspergillosis. Clin Microbiol Rev. 1991; 4:439–456. PMID: 1747861.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download