Abstract
The coexistence of CCND1/IGH and MYC rearrangements in mantle cell lymphoma (MCL) is a rare finding associated with a very poor prognosis. In this study, a patient with blastoid variant (MCL) is reported. The disease was clinically aggressive and refractory to chemotherapy, and the patient only survived for 1 month following diagnosis. Conventional cytogenetic study, FISH, and multicolor FISH (mFISH) demonstrated the involvement of the BCL1/CCND1 locus in a complex translocation, t(3;11)(q25;p15)t(11;14)(q13;q32). In addition, subclonal abnormalities in the 8q24 region, manifested as a t(8;14)(q24;q32)/MYC rearrangement, were identified. To the best of our knowledge, this is the first MCL case in Korea bearing these complex genomic aberrations.
Mantle cell lymphoma (MCL) is a distinct subtype of mature B-cell lymphoma, constituting 5-10% of all non-Hodgkin lymphoma cases. Patients with MCL have a poor prognosis, with a median survival of 3 to 5 yr. The blastoid variant of MCL occurs in 10-20% of all MCL patients, and a more aggressive clinical course has been reported in patients with this variant. MCL is typically positive for B-cell associated markers in addition to bcl-2, cyclin D1, CD5, and/or CD43 and is negative for bcl-6, CD10, and CD23 on immunohistochemistry [1, 2].
The genetic hallmark of MCL, the translocation t(11;14)(q13;q32), juxtaposing the IGH on 14q32 with the BCL1/CCND1 gene on 11q13, results in constitutional overexpression of CCND1 and cell cycle dysregulation, in virtually all cases [3]. Although most CCND1/IGH gene rearrangements result from balanced translocation t(11;14), complex variants of this translocation have been reported, involving additional partner chromosomes leading to various unbalanced translocations [4-6].
It is believed that the overexpression of CCND1 is not, on its own, sufficient to induce malignant transformation of lymphoid cells. Molecular genetic studies have confirmed that additional genomic aberrations are found in the majority of MCL cases, and these aberrations are thought to play a major role in the development and progression of this lymphoma [4, 7, 8]. The 8q24/MYC translocation, t(8;14)(q24;q32), has been described as the driving oncogene in Burkitt's lymphoma (BL). However, abnormalities in MYC have been recognized in other non-Hodgkin lymphomas as well, mainly as a secondary karyotypic change [9]. A few cases of MCL with both t(11;14) and MYC rearrangements have been reported [10]. To the best of our knowledge, coexistence of a complex variant of the t(11;14) and MYC aberrations has not been previously reported in Korea. In this study, we report a CD5-negative blastoid variant of MCL with a rare association of a complex translocation, t(3;11)(q25;p15)t(11;14)(q13;q32) involving CCND1, and subclonal MYC aberrations due to t(8;14)(q24;q32).
An 83-yr-old Korean man was admitted to a university hospital for evaluation of leukocytosis, which had been found during an examination for spinal stenosis. The initial complete blood count results were as follows: hemoglobin level, 10.5 g/dL; platelet count, 176×109/L; and white blood cell count, 125.8×109/L, comprising 5% segmented neutrophils, 7% lymphocytes, 2% monocytes, 2% myelocytes, 3% metamyelocytes, 2% band forms, 3% basophils, 1% atypical lymphocytes, and 75% blast-like lymphoid cells (medium to large-sized with polymorphous nucleus, prominent nucleoli, and a small quantity of basophilic cytoplasm) (Fig. 1). Bone marrow aspiration and biopsy analysis revealed a normocellular marrow with significant infiltration of similar-looking blastoid cells. Immunophenotyping of the bone marrow specimens using flow cytometry demonstrated that the blastoid cells were positive for CD19, CD20, cCD22, CD23, CD79a, HLA-DR, and kappa-restricted surface immunoglobulin, but negative for CD5, CD10, and TdT. Chromosome study revealed 45-47,XY,-9,-11,der(14)t(11;14)(q13;q32),-22,+2~3mar[cp10]/46,XY[2] (Fig. 2A).
Because the der(14)t(11;14) detected by G-banding gave rise to the suspicion that a cryptic t(11;14) might be present, FISH was performed using dual-color dual-fusion BCL1 (11q13) green/IGH (14q32) orange probes (Kreatech Diagnostics, Amsterdam, the Netherlands). The FISH analysis revealed "nuc ish (BCL1×3),(IGH×4),(BCL1 con IGH×2)[97/300]/(BCL1,IGH)×3,(BCL1 con IGH×2)[93/300]/(BCL1×2),(IGH×3),(BCL1 con IGH×1)[31/300]/(BCL1,IGH)×2(BCL1 con IGH×1)[14/300]," showing that more than 60% of the cells harbored 2 BCL1/IGH fused signals representing the 2 derivatives (Fig. 2D). These findings were most compatible with the blastoid variant of MCL. To further characterize the marker chromosomes and identify another submicroscopic CCND1/IGH rearrangement that was undetectable by the G-banding technique, multicolor FISH (mFISH) analysis was performed with a human 24XCyte probe (MetaSystems, Altlussheim, Germany). Hybridization and post-hybridization washes were performed according to the protocols provided by the manufacturers. Image acquisition, processing, and evaluation were performed under a fluorescence microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany) by using Isis/mFISH imaging software (MetaSystems). mFISH analysis revealed derivative chromosomes, der(11)t(3;11)(q25;p15)t(11;14)(q13;q32), der(17)t(8;17)(q?;q?), and der(14)t(8;14)(q24;q32) in 3 metaphase cells (Fig. 2B, C). This result led us to conclude that the 2 marker chromosomes seen in G-banded metaphases were derived from the complex translocation t(11;14) additionally involving chromosome 3 and the translocation between 8q and 17q.
The status of the MYC/IGH rearrangement t(8;14) was confirmed using FISH with a dual-color dual-fusion MYC (8q24) green/IGH (14q32) orange probe (Kreatech Diagnostics). Abnormal patterns with 1 fusion signal were observed in 61% of total cells analyzed. The patient was treated with cytarabine, hydroxyurea, vincristine, and dexamethasone. However, he was refractory to chemotherapy and died 28 days after admission.
The CCND1/IGH gene rearrangement is the most important factor in the diagnosis of MCL, and the microscopic or submicroscopic translocations resulting in complex t(11;14) translocations have only been reported in a few MCL cases [5, 8, 11]. MYC overexpression is significantly associated with negative prognostic influence, more aggressive phenotypes, lower response rates, and shorter overall survival in non-BL patients [9]. The coexistence of complex t(11;14), involving additional partner chromosomes and MYC rearrangement, has been previously reported [5, 8, 11]. Although the presence of multiple chromosome abnormalities in addition to t(11;14) is well known to be associated with blastoid variants and poor prognoses in MCL [12], cases reported with complex t(11;14) and MYC overexpression have tended to show an accelerated course [10, 13-15] (Table 1). The activation of MYC may have a synergistic effect in the MCL progression [16]. In addition, deletions in chromosome 9, consistently detected in this case, are also associated with poor prognosis in MCL [17].
In the present case, conventional cytogenetic analysis provided evidence of 2 marker chromosomes of unknown origin and a translocation t(11;14) (Fig. 2A), and FISH analysis revealed 2 BCL1/IGH fusion signals (Fig. 2D). mFISH analysis was employed to define the discrepancy between the 2 methods, and it revealed that 1 of the marker chromosomes was a derivative chromosome 11, formed by 14q32 fused to 11q13, and the 11p15 end fused to 3q25, which probably caused another fusion signal in the FISH analysis (Fig. 2B). MCL is one of the malignant lymphoid neoplasms with the highest level of clonal heterogeneity, which is indicative of a multistep process of lymphomagenesis [8]. Although every G-banded metaphase cell harbored derivative chromosomes 11 and 14, representing the population showing 2 BCL1/IGH fusion signals in FISH, FISH results showed heterogeneous populations of cells; about 10% of total analyzed cells that showed 1 BCL1/IGH fusion signal and they can be a subclone harboring unbalanced t(11;14). The lack of variation in metaphase G-banding could be due to its lower sensitivity compared with interphase FISH analysis, or to the mitotic error of subclones with unbalanced and more complex karyotypes, or perhaps both.
The mFISH is a powerful tool that facilitates the karyotyping of complex chromosomal rearrangements, cryptic translocations, and marker chromosomes not identifiable by conventional cytogenetics. In this study, mFISH complemented G-banding by identifying several derivative chromosomes. However, it did not clarify the juxtaposition of 14q32 on der(11) and the translocated region of 14qter on chromosome 8 (Fig. 2B). This is attributed to the resolution limit of mFISH, which has been reported to range from 500 to 1,500 kb [18]. The presence of these translocations was completely identified by FISH analyses.
Although the immunophenotype of MCL is relatively characteristic, immunophenotypic heterogeneity in MCL has been described in blastoid variants, including the absence of CD5 and the expression of CD10, BCL6, and CD23 [19, 20]. In conclusion, we report a rare case of MCL, with coexistence of a complex t(11;14) translocation involving CCND1 and MYC aberrations in the blastoid variant of MCL, atypically lacking in CD5 expression. The combination of mFISH and FISH with conventional karyotyping has enabled the elucidation of the compex chromosomal aberrations.
Figures and Tables
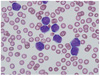 | Fig. 1The peripheral blood smear shows an increased number of medium- to large-sized blastoid cells with polymorphous nuclei and basophilic cytoplasm (Wright-Giemsa stain, ×1,000). |
 | Fig. 2(A) Giemsa-banding karyogram of bone marrow cells: 45-47,XY,-9,-11,der(14)t(11;14)(q13;q32),-22,+2~3mar. M1 and M2 indicate marker chromosome 1 and 2, respectively. (B) Single color galleries of mFISH analysis showing der(14)t(11;14)(q13;q32), der(11) t(3;11)(q25;p15)t(11;14)(q13;q32), der(14)t(8;14)(q24.1;q32), and der(17)t(8;17)(q?;q?) respectively. (C) Diagrammatic representation of der(11)t(3;11)(q25;p15)t(11;14)(q13;q32). The arrows indicate breakpoints in chromosomes 3, 11, and 14. (D) FISH analysis using a dual-color dual-fusion BCL1(green)/IGH(orange) probe, a subclone harboring 2 fusion (F), 1 BCL1(G) and 2 IGH (R) signals. The corresponding chromosomes are marked as F, G, and R on the karyogram (A). |
References
1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 2008. IARC: Lyon.
2. Jares P, Campo E. Advances in the understanding of mantle cell lymphoma. Br J Haematol. 2008. 142:149–165.

3. Bertoni F, Zucca E, Cotter FE. Molecular basis of mantle cell lymphoma. Br J Haematol. 2004. 124:130–140.

4. Salaverria I, Espinet B, Carrio A, Costa D, Astier L, Slotta-Huspenina J, et al. Multiple recurrent chromosomal breakpoints in mantle cell lymphoma revealed by a combination of molecular cytogenetic techniques. Genes Chromosomes Cancer. 2008. 47:1086–1097.

5. Sathanoori M, Shekhter-Levin S, Marks SM, Swerdlow SH. Derivative (14)t(11;14)(q13;q32)t(11;14)(p11.2;p11.2): a novel unbalanced variant of the t(11;14)(q13;q32) translocation in mantle cell lymphoma. Cancer Genet Cytogenet. 2007. 172:158–164.

6. Aamot HV, Tjonnfjord GE, Delabie J, Heim S. Molecular cytogenetic analysis of leukemic mantle cell lymphoma with a cryptic t(11;14). Cancer Genet Cytogenet. 2006. 165:172–175.

7. Salaverria I, Zettl A, Bea S, Moreno V, Valls J, Hartmann E, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J Clin Oncol. 2007. 25:1216–1222.

8. Sander S, Bullinger L, Leupolt E, Benner A, Kienle D, Katzenberger T, et al. Genomic aberrations in mantle cell lymphoma detected by interphase fluorescence in situ hybridization. Incidence and clinicopathological correlations. Haematologica. 2008. 93:680–687.

9. Smith SM, Anastasi J, Cohen KS, Godley LA. The impact of MYC expression in lymphoma biology: beyond Burkitt lymphoma. Blood Cells Mol Dis. 2010. 45:317–323.

10. Reddy K, Ansari-Lari M, Dipasquale B. Blastic mantle cell lymphoma with a Burkitt translocation. Leuk Lymphoma. 2008. 49:740–750.

11. Maravelaki S, Burford A, Wotherspoon A, Joshi R, Matutes E, Catovsky D, et al. Molecular cytogenetic study of a mantle cell lymphoma with a complex translocation involving the CCND1 (11q13) region. Cancer Genet Cytogenet. 2004. 154:67–71.

12. Au WY, Gascoyne RD, Viswanatha DS, Connors JM, Klasa RJ, Horsman DE. Cytogenetic analysis in mantle cell lymphoma: a review of 214 cases. Leuk Lymphoma. 2002. 43:783–791.

13. Michaux L, Wlodarska I, Theate I, Stul M, Scheiff JM, Deneys V, et al. Coexistence of BCL1/CCND1 and CMYC aberrations in blastoid mantle cell lymphoma: a rare finding associated with very poor outcome. Ann Hematol. 2004. 83:578–583.

14. M'Kacher R, Farace F, Bennaceur-Griscelli A, Violot D, Clausse B, Dossou J, et al. Blastoid mantle cell lymphoma: evidence for nonrandom cytogenetic abnormalities additional to t(11;14) and generation of a mouse model. Cancer Genet Cytogenet. 2003. 143:32–38.
15. Au WY, Horsman DE, Viswanatha DS, Connors JM, Klasa RJ, Gascoyne RD. 8q24 translocations in blastic transformation of mantle cell lymphoma. Haematologica. 2000. 85:1225–1227.
16. Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E, et al. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood. 2002. 99:4554–4561.

17. Dreyling M, Hoster E, Bea S, Hartmann E, Horn H, Hutter G, et al. Update on the molecular pathogenesis and clinical treatment of Mantle Cell Lymphoma (MCL): minutes of the 9th European MCL Network conference. Leuk Lymphoma. 2010. 51:1612–1622.

18. Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Singhal S, Mehta J, et al. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood. 1998. 92:4269–4278.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download