Abstract
In July 2010, we identified an outbreak of vancomycin-resistant enterococci (VRE) in our 26-bed neonatal intensive care unit. We performed an epidemiological investigation after clinical cultures of 2 neonates were positive for VRE. Identification, susceptibility testing, and molecular characterization were performed. Cultures of 3 surveillance stool samples of inpatients and 5 environmental samples were positive for VRE. All isolates were identified as Enterococcus faecium containing the vanA gene. Two distinct clones were identified by performing pulsed-field gel electrophoresis. The 2 clones exhibited different pulsotypes, but they represented identical Tn1546 types. Two sequence types, ST18 and ST192, were identified among all of the isolates with multilocus sequence typing. Our investigation determined that the outbreak in the neonatal intensive care unit was caused by 2 genetically different clones. The outbreak may have occurred through clonal spread and horizontal transfer of the van gene.
Vancomycin-resistant enterococci (VRE), which have been established as important nosocomial pathogens worldwide [1], have become endemic in a number of hospitals. VRE colonization of the gastrointestinal tract, which usually precedes infection, may persist for long periods and serve as silent reservoirs for colonization in other patients [2, 3]. Dissemination of vancomycin resistance can occur through both clonal expansion of VRE and horizontal transfer of van genes to other bacteria [4]. Currently, multiple clones of vancomycin resistance genes and their horizontal transmission are commonly encountered. VRE infection among neonates appears to be quite rare because external contact is limited due to the restricted access to neonatal intensive care units (NICU). Therefore, the major cause of VRE transmission in the NICU is the indirect spread of VRE by health care personnel or through contaminated environments. Asymptomatic patients with VRE colonization can serve as continuing reservoirs because healthcare workers may not adequately wash their hands between contacts with different neonates. Risk factors for VRE colonization in neonates include low birth weight, prematurity, and long-term antimicrobial therapy [5]. Most neonatal VRE infections have been associated with clusters and outbreaks [5-8].
Since the first isolation of VRE in our institution in 1996, the prevalence of VRE has dramatically increased. In the NICU, however, VRE infections have been rarely reported, and all of them were sporadic cases until 2010. We describe, herein, a VRE outbreak that was caused by 2 genetically different clones in the NICU and the clinical presentations, the interventions used to control the spread of VRE, and the molecular epidemiologic investigation
This study was conducted in a 1,000-bed university hospital. The NICU ward has 2 rooms with 26 beds, as shown in Fig. 1. The NICU is staffed by 1 pediatrician, 5 residents, and 29 nurses who provide care for critically ill patients of age less than or equal to 6 months. All healthcare workers practice standard precautions. In July 2010, VRE was isolated from a sputum sample of a premature neonate (Index Case 1). Subsequently, VRE was isolated from a urine culture of another neonate (Index Case 2).
A 780-g baby boy, who was the first born of a set of twins, was born by normal spontaneous vaginal delivery (NSVD) to a G1P0 21-yr-old mother at 25 weeks of gestation. Apgar scores were 2 and 3 at 1 and 5 min, respectively. After birth, he received surfactant treatment because he had developed a number of complications of prematurity such as respiratory distress requiring mechanical ventilation and oxygen therapy. He was administered ampicillin-sulbactam until his primary cultures were negative. On the 5th day in the NICU, he developed abdominal distension, and an X-ray radiograph of his abdomen showed the ileus sign indicating necrotizing enterocolitis (NEC). On the 8th day, he underwent small bowel resection for bowel perforation, and on the 15th day, he developed pulmonary hemorrhage and intraventricular hemorrhage. On the 31st day, a sputum sample was obtained, and the sputum culture grew VRE. Ampicillin-sulbactam was replaced with ceftriaxone and linezolid. Although the sputum culture stopped growing VRE on the 36th day, VRE continued growing in cultures obtained from rectal swab samples. On the 77th day, he started feeding training, but bile regurgitation was observed, and bowel obstruction was suspected. He underwent exploratory laparatomy, and small bowel resection was performed. On the 135th day, feeding training was restarted, but bile regurgitation was observed again. An imaging study showed that the NEC had aggravated, and metabolic acidosis and pneumonia had developed. On the 142nd day, he died due to multiple organ failure and sepsis. Stool cultures remained positive for VRE throughout his hospitalization.
A 2,300-g baby boy was born to a G1P0 27-yr-old mother at 36 weeks of gestation by NSVD. Apgar scores were 8 and 9 at 1 and 5 min, respectively. His health conditions during the first week were uneventful. On the 7th day after his birth, he showed lethargy and poor oral intake. He was diagnosed with aseptic meningitis, disseminated intravascular coagulation, and neonatal jaundice on the basis of initial laboratory test results. He was admitted to the NICU and received vancomycin and ceftriaxone for 7 days. On the 6th day of his hospital stay, he developed bradycardia and exhibited increased cardiac enzyme levels with an ejection fraction of 19%, which resulted in the diagnosis of myocarditis. Urine and rectal swab samples were collected on the 18th and 23rd day, and cultures of these samples grew VRE. The patient initially received vancomycin and ceftriaxone; these were then replaced with ampicillin-sulbactam. Blood cultures and urine cultures were negative after 1 week of ampicillin-sulbactam treatment and for 1 week after discontinuation of the ampicillin-sulbactam treatment. The patient's clinical condition improved, and no complications were observed. On the 28th day, the patient was discharged, and echocardiography performed at 2 months later revealed a normal ejection fraction (61%).
Immediately after the occurrence of the second case of VRE, a special team was formed to coordinate the management of the outbreak. We performed active surveillance cultures for VRE on samples from all of the 26 NICU patients, 42 health care workers, and their environments. Environmental samples were collected on swabs moistened with sterile saline from multiple sites in the NICU. All patients who were identified with VRE were placed in strict cohorting. Infection control measures were strengthened, and all patient areas were thoroughly cleaned. Screening was repeated every week until VRE-positive patients were present in the NICU.
Organisms were identified by conventional biochemical reactions with the Vitek identification system (bioMérieux, Inc., Hazelwood, MO, USA) and with the API Strep system (bioMérieux, Inc.). All isolates were typed by performing pulsed-field gel electrophoresis (PFGE) on a CHEF-DR III apparatus (Bio-Rad Laboratories, Inc., Richmond, VA, USA). After digestion with SmaI, genomic DNA was separated by performing electrophoresis with ramped pulse times of 5-30 sec at 6 V/cm for 19 hr. The banding patterns were interpreted according to Tenover et al. [9]. The vancomycin resistance genotypes were determined by performing PCR with primers that were specific for the sequences of the vanA, vanB, vanC1, and vanC2/C3 genes, as described previously [10].
Isolates were characterized by their Tn1546 elements. DNA was extracted with the QIAGEN DNeasy Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instruction. For the structural analysis of Tn1546, overlapping PCR amplification of the internal regions of the elements was performed, as described previously [11]. The purified PCR products were directly sequenced on an ABI Prism 3100 DNA SEQUENCER (Applied Biosystems, Foster City, CA, USA) and analyzed using DNASIS for Windows v.2.6 (Hitachi Solutions America, Ltd., South San Francisco, CA, USA).
Multilocus sequence typing (MLST) was performed according to the protocol previously described [12]. Internal fragments of 7 housekeeping genes (adk, atpA, ddl, gdh, gyd, purK, and pstS) were amplified by PCR and directly sequenced. The allele number for each gene was assigned according to the Enterococcus faecium MLST database (http://www.mlst.net). The combination of the allelic sequences for the 7 genes yielded the allelic profile or sequence type (ST) for each isolate.
Cultures of 6 sputum samples of index case 1 and 2 and urine samples of index case 2 were positive for VRE. Cultures of 3 surveillance stool samples of inpatients and 5 environmental samples were positive for VRE. All surveillance cultures of medical staff were negative for VRE. Culture-positive samples from the environment were obtained from the door handle of isolation room 1, the shelves of storage room 2, and the tables of the office and treatment room (Fig. 1). The strengthened infection control measures were terminated after 2 months during which no additional new cases were detected. All subsequent environmental VRE cultures were negative.
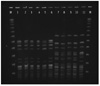
All isolated VREs were E. faecium, and PCR analysis showed that the resistance gene was vanA. PFGE, MLST, and the structural analysis of Tn1546 were performed for 16 isolates. The obtained results are summarized in Table 1. Using PFGE, we identified 3 clones of VRE: clone A, clone B1, and clone B2 (Fig. 2). Clones B1 and B2 had 4 band differences and were included in the genetically related category. Of the 16 isolates, 11 belonged to clone A, 4 belonged to clone B1, and 1 belonged to clone B2. Six isolates obtained from index case 1 exhibited identical PFGE profiles (clone A). However, the PFGE patterns of the 2 isolates obtained from index case 2 were different from one another; the first isolate belonged to clone A, and the second isolate belonged to clone B1.
Genotyping of all of the isolates by MLST revealed 2 different STs; of the 16 isolates, 11 were ST18 and 5 were ST192. The structural analysis of Tn1546 showed that all isolates had identical Tn1546 type. The type represented 2 copies of IS1216V at the left end of Tn1546 and in the vanX-vanY intergenic region as well as IS1542 in the orf2-vanR intergenic region.
Our results show that the VRE outbreak in the NICU was caused by 2 genetically different VRE clones. The index case 1 was identified on July 7, 2009, and Clone A was responsible for index case 2, VRE carriage (as detected in 1 case) and environmental contamination (as detected in 3 samples). The index case 2 was identified on July 9, 2009, and clone B1 was responsible for VRE carriage (as detected in 2 cases) and environmental contamination (as detected in 2 samples).
There were 2 predominant clones in the NICU. These 2 clones had different PFGE profiles but identical Tn1546 type. Two different STs, namely, ST18 and ST192 were identified among all isolates. ST18 and ST192 are double-locus variants of ST17, which was the founder of the clonal complex 17 (CC17). Most nosocomial E. faecium strain types that are associated with outbreaks belong to a single clonal complex called the CC17 lineage [13-15]. CC17 dissemination appears to have preceded the emergence of vancomycin resistance, as shown by its predominance in hospitals throughout the world [16, 17].
Our results suggested that the outbreak occurred through clonal spread and horizontal transfer of the van gene. At first, the dissemination of VRE from index case 1 to index case 2 occurred through clonal expansion. While index case 2 harbored VRE acquired from index case 1, horizontal transfer of Tn1546 to other E. faecium occurred in the gastrointestinal tract of index case 2. Because infant-to-infant spread of VRE did not occur, VRE might have been transmitted indirectly through contact with health care personnel or through contaminated environment. Index case 1 carried VRE in his intestine throughout his hospitalization. However, after intervention, no new VRE cases were detected, and all subsequent environmental samples were negative for VRE. Our results suggest that VRE spread can be prevented by adopting intensive infection control measures, even if VRE carriers are present. Once VRE is colonized, the clearance of carriers is difficult to achieve. There is currently no effective VRE decolonization regimen. However, enforcing cleaning measures and encouraging hand hygiene and strict cohorting can prevent the spread of VRE in hospitals. Early interventions are associated with a high likelihood of controlling hospital outbreak. Thus, at the onset of an outbreak, the possibility of environmental contamination should be considered, and measures to tackle environmental contamination should be adopted.
Figures and Tables
Fig. 1
Map of the neonatal intensive care unit showing the location of the patients with vancomycin-resistant enterococci (VRE). Numbered rectangles, location of beds inside the room; Gray rectangles, environmental locations that showed presence of VRE.

Fig. 2
Pulsed-field gel electrophoresis of vancomycin-resistant isolates. M, molecular weight marker; lane 1, isolate from index case 1; lane 2, isolate from index case 2; lane 3, isolate from contact patient 1; lanes 4 to 6, isolates from environment; lane 7, isolate from index case 2; lane 8, isolate from contact patient 2; lanes 9 and 10, isolates from environment; lane 11, isolate from contact patient 3.
References
1. Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000. 13:686–707.

2. Lee WG, Park IJ, Jin HY, Park MH. Relapse and reacquisition of rectal colonization by vancomycin-resistant Enterococcus faecium after decolonization. Epidemiol Infect. 2010. 138:1449–1453.
3. Montecalvo MA, de Lencastre H, Carraher M, Gedris C, Chung M, VanHorn K, et al. Natural history of colonization with vancomycin-resistant Enterococcus faecium. Infect Control Hosp Epidemiol. 1995. 16:680–685.
4. Gholizadeh Y, Courvalin P. Acquired and intrinsic glycopeptide resistance in enterococci. Int J Antimicrob Agents. 2000. 16:S1. S11–S17.

5. Sherer CR, Sprague BM, Campos JM, Nambiar S, Temple R, Short B, et al. Characterizing vancomycin-resistant enterococci in neonatal intensive care. Emerg Infect Dis. 2005. 11:1470–1472.

6. Singh N, Léger MM, Campbell J, Short B, Campos JM. Control of vancomycin-resistant enterococci in the neonatal intensive care unit. Infect Control Hosp Epidemiol. 2005. 26:646–649.

7. Ergaz Z, Arad I, Bar-Oz B, Peleg O, Benenson S, Minster N, et al. Elimination of vancomycin-resistant enterococci from a neonatal intensive care unit following an outbreak. J Hosp Infect. 2010. 74:370–376.

8. Yüce A, Karaman M, Gülay Z, Yulug N. Vancomycin-resistant enterococci in neonates. Scand J Infect Dis. 2001. 33:803–805.
9. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–2239.

10. Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995. 33:1434.

11. Woodford N, Adebiyi AM, Palepou MF, Cookson BD. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998. 42:502–508.

12. Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, et al. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002. 40:1963–1971.
13. Freitas AR, Tedim AP, Novais C, Ruiz-Garbajosa P, Werner G, Laverde-Gomez JA, et al. Global spread of the hyl(Efm) colonization-virulence gene in megaplasmids of the Entercoccus faecium CC17 polyclonal subcluster. Antimicrob Agents Chemother. 2010. 54:2660–2665.
14. Galloway-Peña JR, Nallapareddy SR, Arias CA, Elipoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009. 200:1566–1573.
15. Bonora MG, Olioso D, Lo Cascio G, Fontana R. Phylogenetic analysis of vancomycin-resistant Enterococcus faecium genotypes associated with outbreaks or sporadic infections in Italy. Microb Drug Resist. 2007. 13:171–177.
16. Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006. 9:454–460.

17. Klare I, Konstabel C, Mueller-Bertling S, Werner G, Strommenger B, Kettlitz C, et al. Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur J Clin Microbiol Infect Dis. 2005. 24:815–825.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download