Abstract
Rapid and accurate detection of norovirus is essential for the prevention and control of norovirus outbreaks. This study compared the effectiveness of a new immunochromatographic assay kit (SD BIOLINE Norovirus; Standard Diagnostics, Korea) and real-time reverse transcription-PCR (RT-PCR) for detecting norovirus in fecal specimens. Compared with real-time RT-PCR, the new assay had sensitivity, specificity, positive predictive value, and negative predictive value of 76.5% (52/68), 99.7% (342/343), 98.1% (52/53), and 95.5% (342/358), respectively. The sensitivity of the assay was 81.8% (18/22) for GII.3 and 75.7% (28/37) for GII.4. None of the 38 enteric virus-positive specimens (3 for astrovirus, 5 for enteric adenovirus, and 30 for rotavirus) tested positive in the cross-reactivity test performed by using this assay. The new immunochromatographic assay may be a useful screening tool for the rapid detection of norovirus in sporadic and outbreak cases; however, negative results may require confirmatory assays of greater sensitivity.
Noroviruses, which belong to the family Caliciviridae, are small non-enveloped RNA viruses that possess a linear, positive-sense, and single-stranded RNA genome. Noroviruses are genetically classified into 5 groups (GI V); GI, GII, and GIV cause human infections [1, 2]. Noroviruses are the most common cause of epidemic gastroenteritis, accounting for more than 23 million cases of gastroenteritis annually in the United States and about 50% of all cases of outbreaks worldwide, and they are a significant cause of sporadic cases of community-related gastroenteritis [3-5]. Norovirus spreads easily because of several characteristics such as a low infectious dose (as few as 10-100 particles), large quantities of the virus in feces and vomit, and environmental stability [3, 4]. Thus, the rapid and accurate detection of norovirus is essential for the prevention and control of norovirus outbreaks.
Methods used for the detection of norovirus in clinical specimens include electron microscopy of fecal specimens, reverse transcription PCR (RT-PCR), real-time RT-PCR, enzyme immunoassays (EIAs), and immunochromatographic assays. An immunochromatographic assay is rapid, providing a result within 30 min (usually between 15 and 30 min), but some assays have inadequate sensitivity for diagnosis of sporadic norovirus disease. Recently, a new immunochromatographic assay kit (SD BIOLINE Norovirus; Standard Diagnostics, Yongin, Korea) has been developed for the rapid detection of norovirus in fecal specimens. This study compared the effectiveness of this new immunochromatographic test kit and real-time RT-PCR assay for detecting norovirus in fecal specimens.
A total of 411 fecal specimens from patients (inpatients and outpatients) presenting with symptoms of acute gastroenteritis were obtained between December 2010 and February 2011. The ages of the patients ranged from 5 weeks to 94 yr (average, 22.6 yr). For the new immunochromatographic assay, stool suspensions were prepared in 1 mL of a dilution buffer supplied in the kit; the assay was conducted according to the manufacturer's instructions. For the molecular assay, each fecal specimen was diluted to yield a 10% suspension in phosphate-buffered saline and was clarified by centrifugation at 8,000×g for 15 min. The supernatants were collected and stored at -80℃ until use. Viral RNA was extracted from 150 µL of each fecal supernatant by using a viral nucleic acid preparation kit (Greenmate Biotech, Seoul, Korea), in accordance with the manufacturer's instructions. The extracted RNA was dissolved in 50 µL of nuclease-free water and stored at -80℃ until it was used for real-time and semi-nested RT-PCR.
Real-time RT-PCR was conducted using an AccuPower® Norovirus Real-Time RT-PCR Kit (Bioneer, Daejeon, Korea) in accordance with the manufacturer's instructions; the 50-µL reaction mixture contained 10 µL of RNA and each primer at a final concentration of 0.3 µM. Reverse transcription, amplification, and detection were performed using Exicycler™ 96 (Bioneer) under the following conditions: initial hold at 45℃ for 15 min and 95℃ for 5 min, followed by 45 cycles at 95℃ for 5 sec, 55℃ for 5 sec, and 25℃ for 1 min. Positive and negative control reactions were included in each run. A sample with threshold cycle (Ct) value <35 and a typical sigmoid curve was defined as positive. To determine the presence of PCR inhibitors, a mixture containing 5 µL of clarified fecal extract of the test specimen, 5 µL of clarified fecal extract of a known norovirus-positive specimen, and 130 µL of nuclease-free water was prepared, and the norovirus real-time RT-PCR was then carried out as described above. A negative real-time RT-PCR result was considered as evidence of the presence of PCR inhibitors.
In an effort to identify norovirus genotypes, we performed direct sequencing of all samples that tested positive for norovirus by the real-time RT-PCR assay. For sequencing, semi-nested RT-PCR was conducted as described previously [6]. The products were purified using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA). The nucleotide sequences were determined using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems Foster City, CA, USA) with GI-R1M and GII-R1M primers in the 3730 XL DNA Analyzer (Applied Biosystems). A BLAST search of GenBank sequences was conducted to determine norovirus genotypes.
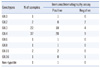
Of the 411 fecal specimens, 68 tested positive by the real-time RT-PCR assay and 53 tested positive by the new assay. Among the PCR-negative specimens, none showed inhibition of PCR. Compared with real-time PCR, the new assay had sensitivity, specificity, positive predictive value, and negative predictive value of 76.5% (52/68), 99.7% (342/343), 98.1% (52/53), and 95.5% (342/358), respectively (Table 1). The new assay detected the GII.1, GII.3, GII.4, GII.6, GII.8, and GII.11 genotypes, but not the GII.2 and GII.16 genotypes (Table 2). The sensitivity of the assay was 81.8% (18/22) for GII.3 and 75.7% (28/37) for GII.4.
To evaluate the cross-reactivity of the new assay, we tested 343 norovirus PCR-negative specimens by using real-time RT-PCR for other enteric viruses (astrovirus, enteric adenovirus, and rotavirus). None of the other 38 enteric virus-positive specimens (3 for astrovirus, 5 for enteric adenovirus, and 30 for rotavirus) tested positive by the new assay. For evaluation of the detection limits of the new assay, we prepared 5-fold serial dilutions of 6 fecal specimens (3 GII.3 and 3 GII.4 isolates), and each dilution was tested by using the new assay and real-time RT-PCR. Standard curves were generated using reference norovirus RNA transcripts (Bioneer). The number of RNA copies in a sample was determined on the basis of the CT value, and the corresponding numbers of genomic copies were extrapolated from the appropriate standard curve. The average lowest number of copies of GII.3 and GII.4 that could be detected by the new assay were 1.7×107 and 9.5×107 RNA copies/g feces, respectively.
Human noroviruses were first discovered by using electron microscopy. However, the usefulness of electron microscopy in clinical settings is limited because of the low sensitivity and time-consuming nature of the technique, as well as the requirement for expensive equipment. Since the molecular cloning of the Norwalk virus genome in 1990 and the introduction of PCR as a diagnostic tool, molecular assays such as RT-PCR and real-time RT-PCR have been developed for the detection of norovirus [1]. Commercially available viral-antigen detection assays, which have been developed on the basis of the principle of EIA and immunochromatography, are easily available and can be used for rapid detection. EIA assays for detecting norovirus are highly specific, but vary in sensitivity from 32% to 90% [7, 8]. When a real-time RT-PCR assay was used as the gold standard test for comparison, the sensitivity of the immunochromatographic assays ranged from 57% to 82%, and the specificity ranged from 98% to 100% [9-11]. However, when a conventional RT-PCR assay was used as the standard test for comparison, the sensitivity of the immunochromatographic assays ranged from 70% to 79%, and the specificity ranged from 94% to 100% [8, 12, 13]. Although some antigen detection tests have inadequate sensitivity for detecting sporadic cases, assays with high specificity may be useful to rapidly detect norovirus during outbreaks.
The sensitivity of the new assay was comparable to that of the other immunochromatographic assays. Moreover, the results of the assay showed very high specificity for the detection of norovirus and no cross-reactivity for other enteric viruses. The sensitivity of EIA and immunochromatographic assays is often genotype-dependent [4, 7]. The new assay displayed good sensitivity for a wide range of GII genotypes, except for GII.2 and GII.16; however, the number of samples containing some norovirus genotypes was too small to indicate statistical significance. Since the ability of the new assay to detect GI genotypes was not evaluated in this study, further studies are needed to evaluate the ability of this assay to detect GI genotypes.
In conclusion, the new immunochromatographic assay may be a useful screening tool for the rapid detection of norovirus in sporadic and outbreak cases; however, a negative result may require confirmatory assays of greater sensitivity.
Figures and Tables
Acknowledgements
We thank the Standard Diagnostics Company (Yongin-si, Korea) for kindly providing the SD BIOLINE Norovirus immunochromatography kit for this study.
References
1. Costantini V, Grenz L, Fritzinger A, Lewis D, Biggs C, Hale A, et al. Diagnostic accuracy and analytical sensitivity of IDEIA Norovirus assay for routine screening of human norovirus. J Clin Microbiol. 2010. 48:2770–2778.

2. Green KY. Knipe DM, Howley PM, editors. Caliciviridae: The Noroviruses. Fields virology. 2007. 5th ed. Philadelphia: Lippincott Williams and Wilkins;949–979.
4. Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol. 2009. 44:1–8.

5. Boga JA, Melón S, Nicieza I, De Diego I, Villar M, Parra F, et al. Etiology of sporadic cases of pediatric acute gastroenteritis in asturias, Spain, and genotyping and characterization of norovirus strains involved. J Clin Microbiol. 2004. 42:2668–2674.

6. Park KS, Jeong HS, Baek KA, Lee CG, Park SM, Park JS, et al. Genetic analysis of norovirus GII.4 variants circulating in Korea in 2008. Arch Virol. 2010. 155:635–641.

7. Gray JJ, Kohli E, Ruggeri FM, Vennema H, Sánchez-Fauquier A, Schreier E, et al. European multicenter evaluation of commercial enzyme immunoassays for detecting norovirus antigen in fecal samples. Clin Vaccine Immunol. 2007. 14:1349–1355.

8. Khamrin P, Nguyen TA, Phan TG, Satou K, Masuoka Y, Okitsu S, et al. Evaluation of immunochromatography and commercial enzyme-linked immunosorbent assay for rapid detection of norovirus antigen in stool samples. J Virol Methods. 2008. 147:360–363.

9. Bruins MJ, Wolfhagen MJ, Schirm J, Ruijs GJ. Evaluation of a rapid immunochromatographic test for the detection of norovirus in stool samples. Eur J Clin Microbiol Infect Dis. 2010. 29:741–743.

10. Derrington P, Schreiber F, Day S, Curtis C, Lyon M. Norovirus Ridaquick: a new test for rapid diagnosis of norovirus. Pathology. 2009. 41:687–688.

11. Kirby A, Gurgel RQ, Dove W, Vieira SC, Cunliffe NA, Cuevas LE. An evaluation of the RIDASCREEN and IDEIA enzyme immunoassays and the RIDAQUICK immunochromatographic test for the detection of norovirus in faecal specimens. J Clin Virol. 2010. 49:254–257.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download