Abstract
Background
Introduction of the Luminex panel reactive antibody (PRA)-single antigen (SA) assay has increased the detection rates of unacceptable antigens in sensitized patients; the calculated PRA (CPRA) level represents the percentage of actual organ donors that express 1 or more of these unacceptable antigens. We developed a CPRA calculator based on the HLA frequencies in Koreans to measure sensitization levels in Korean patients.
Methods
To develop the calculator, we obtained the HLA-A, HLA-B, and HLA-DR phenotypes of 1,622 Koreans, and compared these with previously reported frequencies in Koreans. Sera from patients awaiting kidney transplantation were tested for HLA antibodies by Luminex PRA-screen, PRA-identification (ID), and PRA-SA assays. The measured %PRA from the PRA-screen (N=55) and PRA-ID (N=71) were compared to the %CPRA for the unacceptable antigens obtained from PRA-SA.
Results
Phenotype frequencies used for the CPRA calculator agreed with previously reported data. The concordance rates among the 3 PRA methods for the detection of class I and class II antibodies were 76.1-81.8% (kappa, 0.519-0.636) and 72.7-83.6% (0.463-0.650), respectively. For the detection of broadly sensitized sera (>50% or >80%), the concordance rates were over 80%. In sera with 80-100% CPRA, 91.7% and 94.4% of the samples had concordant results (80-100% PRA) in the PRA-screen and PRA-ID assay, respectively.
Go to : 
Sensitized patients waiting for renal allografts that have preformed antibodies against donor-specific HLA antigens are at risk of hyperacute, accelerated acute antibody-mediated rejection and poor graft outcome. Panel reactive antibodies (PRAs) have been used to measure the relative degree of sensitization in renal allograft recipients. PRA levels represent the percentage of likely cross-match incompatible donors, and are determined by testing recipient sera against cells from a panel of HLA-typed donors or solubilized HLA antigens attached to a solid phase. The panels should be representative of the local pools of potential organ donors. However, the results of PRA testing can be highly variable and inconsistent depending on the panel composition and the techniques used for HLA antibody detection [1, 2].
The development of solid phase-based assays that use solubilized HLA antigens has greatly increased the ability to detect and identify HLA-specific antibodies [3-5]. In particular, the use of recombinant single antigens (SA) in the Luminex assay makes it possible to detect HLA-specific antibodies with greater sensitivity and accuracy. Calculated panel reactive antibody (CPRA) values are based on the HLA antigens that are listed as unacceptable for renal transplant candidates. The unacceptable HLA antigens can be identified by the presence of HLA antibodies in the sera of transplant recipients [2]. This assessment can predict crossmatch-positive donor kidneys (as a virtual crossmatch) and has increased the efficiency of organ allocation.
A kidney allocation process using CPRA has been established in the United Network for Organ Sharing (UNOS) and Eurotransplant allocation system. UNOS awards sensitized patients with CPRA levels ≥80 an additional point to increase their access to potentially compatible donors. Furthermore, the organ procurement network does not offer organs expressing unacceptable HLA antigens to recipients who have HLA antibodies against those particular antigens.
In contrast, the Korean Network for Organ Sharing (KONOS) does not administer the PRA or CPRA, and only uses crossmatch results to measure sensitization for the renal allocation system. This is probably due to the variability in PRA methods, a lack of organized guidelines, and the differences between the antigen composition in commercial PRA panels and that in the Korean population. Therefore, a more uniform and accountable method for measuring sensitization to HLA antigens based on Korean HLA phenotypes is needed. In this study, we developed a CPRA calculator using the HLA phenotypes of Koreans to represent the percentage of actual donors expressing unacceptable HLA antigens; then, we compared this CPRA approach with the traditional PRA approach using Luminex technology.
Go to : 
We developed a "Catholic Medical Center (CMC)-CPRA calculator" with Microsoft Excel using HLA phenotypes derived from 1,662 healthy Korean donors who underwent HLA-A, HLA-B, and HLA-DR typing at Seoul St. Mary's Hospital from May 2005 to March 2010 for related or unrelated organ donation. HLA phenotypes were determined by a molecular typing method using PCR-sequence specific oligonucleotides (Dynal RELI HLA-A, -B, and DRB kits; Dynal Biotech LTD, Wirral, UK). The HLA typing results were validated whether observed genotype frequencies were consistent with Hardy-Weinberg equilibrium. When HLA-A, HLA-B, or HLA-DR antibodies detected by Luminex PRA-SA testing were entered into the CPRA calculator, a CPRA value (%CPRA) was automatically determined as the percentage of persons with unacceptable antigens (Fig. 1). We compared the HLA phenotype frequencies from this CMC-CPRA calculator to those in previous reports based on Korean populations [6, 7]. The HLA phenotype frequencies used for the CMC-CPRA calculator were also compared to those for the UNOS- and Eurotransplant Reference Laboratory (ETRL)-calculators on the websites at http://optn.transplant.hrsa.gov and http://www.etrl.org/etrlpra, respectively.
Seventy-one serum specimens obtained from patients on the waiting list for kidney transplantation were tested by the PRA-SA assay using LIFECODES LSA class I and class II kits (Gen-Probe Transplant Diagnostics Inc., Stamford, CT, USA), and HLA class I and class II specificities were identified according to the manufacturer's instructions. SA beads contained over 90 different recombinant HLA-A, HLA-B, and HLA-C class I antigens, and over 60 recombinant HLA-DRB, HLA-DQB, and HLA-DPB class II antigens. A bead was considered positive if 2 or more of the adjusted values were above the 1,000 median fluorescence index (MFI) cutoff on the Luminex 200 platform (Luminex Corp., Austin, TX, USA). To determine the CPRA value, the detected HLA-A, HLA-B, and HLA-DR antibody specificities were entered into the CMC-CPRA calculator.
Out of the 71 serum samples, 55 were tested in a PRA screen using the LIFECODES LifeScreen Deluxe kit (Gen-Probe Transplant Diagnostics Inc.). Sera with discrepant results in the PRA-screen and PRA-ID assays were retested after SeraClean treatment to reduce nonspecific reactions. LifeScreen Deluxe is a qualitative Luminex assay that contains 6 different Class I CREG- and 4 different Class II CREG-enriched beads in addition to beads coated with each class I and class II pooled antigen. If at least 1 of the 7 class I HLA beads or at least 1 of the 5class II HLA beads was positive, the sample was considered positive for class I or class II HLA-specific antibodies, respectively. The PRA value (%PRA) for the PRA-screen test was calculated by dividing the number of positive bead reactions by the number of CREG beads (6 for class I, and 4 for class II).
All 71 sera were tested for PRA-ID using LIFECODES Class I and Class II ID kits (Gen-Probe Transplant Diagnostics Inc.), and the %PRAs for the PRA-ID test were calculated according to the manufacturer's instructions as the percentage of positive bead reactions among the 50 class I beads and 42 class II beads. To compare these findings with the CPRA values, which combined class I and class II specificities, the greater %PRA for either the class I or class II values was selected.
Each locus was tested for Hardy-Weinberg equilibrium using the GENEPOP program, and exact P values were estimated by the Markov chain method [8, 9]. Statistical analyses were performed using SPSS version 12.0 (SPSS, Chicago, IL, USA). Chisquare analysis was used to compare the phenotype frequencies from different studies. Agreement between the CPRA and PRA values was assessed according to kappa coefficient (0.001-0.2 indicates slight concurrence, 0.201-0.4 indicates fair agreement, 0.401-0.6 shows moderate agreement, 0.601-0.8 indicates substantial concurrence, and 0.801-0.999 shows excellent agreement).
Go to : 
In the CMC-CPRA calculator, the genotype frequencies for the HLA-A, HLA-B, and HLA-DR loci were in Hardy-Weinberg equilibrium (P =0.825, 0.477, and 0.557, respectively). When we compared the phenotype frequencies used for the CMC-CPRA calculator with those obtained from 2 studies of Korean populations [6, 7], the B35, B62, and DR15 antigens had greater than a 2 percent difference compared to both of the previous studies (Table 1). Several antigens, including A11, B35, B44, B46, B56, B62, B63, B75, DR11, DR12, DR15, and DR16 differed significantly from either one or both of previous data (P <0.05). When we compared the phenotype frequencies from the CMC-, UNOS-, and ETRL-CPRA calculators, the frequencies of 11 antigens (A24, A33, B46, B54, B58, B61, DR4, DR8, DR9, DR12, and DR14) were much higher (>10% difference) in the CMC-CPRA calculator than in the UNOS- or ETRL-CPRA calculators. The frequencies of 6 antigens (A1, A3, B7, B8, DR3, and DR11) in the CMC-CPRA calculator were much lower (>10% difference) than those in the UNOS- or ETLR-CPRA calculators.
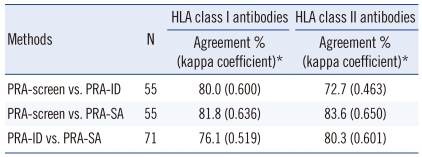
Agreement between the PRA-screen, PRA-ID, and PRA-SA tests when the results represent either the presence or absence of HLA antibodies is shown in Table 2. For the detection of class I HLA-specific antibodies, the PRA-screen had 80.0% and 81.8% agreement with the PRA-ID and PRA-SA methods, respectively. The PRA-ID and PRA-SA tests had moderate agreement (76.1%; kappa coefficient, 0.519) for the detection of class I HLA-specific antibodies. For the detection of class II antibodies, the PRA-screen test had moderate agreement (72.7%; kappa coefficient, 0.463) with the PRA-ID due to low co-negativity (58.6%). When the results were represented as either positive or negative with an 80% cut-off, the PRA-SA tests had 83.6% (0.618) and 81.7% (0.597) agreement with the PRA-screen and PRA-ID, respectively. With a 50% cut-off for the detection of highly sensitized sera, the assays had 81.8% (0.631) (PRA-screen vs. PRA-SA) and 83.1% (0.663) (PRA-ID vs. PRA-SA) agreement.
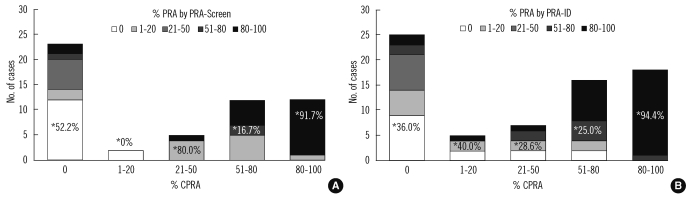
We compared the PRA values from the PRA-screen and PRA-ID tests to the CPRA values calculated from the PRA-SA result using CMC-PRA calculator. Fig. 2 shows the distribution of PRA results within the 5 groups of CPRA levels. In the 80-100% CPRA group, the PRA values from the PRA-screen and PRA-ID had 91.7% and 94.4% agreement with the CPRA values, respectively. However, in the groups with 0%, 1-20%, 21-50%, or 51-80% CPRA, the concordance rates were less than 80%. There were a considerable number of cases with higher %PRA than %CPRA in the lower CPRA groups.
Go to : 
The use of CPRA is rapidly being adopted by transplant laboratories worldwide. The UNOS established a CPRA calculator using HLA-A, HLA-B, HLA-DR, and HLA-DQ frequencies derived from the phenotypes of more than 12,000 donors recently entered into the OPTN registry [10]. The ETRL-CPRA program uses HLA typing data from about 4,000 organ donors, including 1,000 donors from different participating countries. The ETRL-CPRA calculation includes the frequencies of HLA-A, HLA-B, HLA-C, HLA-DR, and HLA-DQ.
In clinical laboratories in Korea, the use of PRA tests with a solid phase assay is increasing. Additionally, PRA-SA tests have recently been introduced in a few laboratories. Therefore, virtual crossmatch prediction by CPRA using unacceptable antigens could be used to increase the kidney allocation efficiency for sensitized patients awaiting transplantation.
To obtain the CPRA, a list of unacceptable antigens is required instead of a PRA value. In this study, we developed a CMC calculator based on the HLA-A, HLA-B, and HLA-DR frequencies derived from the phenotypes of 1,662 Korean donors. The CPRA represents the percentage of potential donors who have 1 or more unacceptable HLA-A, HLA-B, or HLA-DR antigens. Although the pool of HLA phenotypes for the CMC-CPRA calculator was much smaller than those for the UNOS or ETRL calculators, the phenotype data from the CMC-CPRA calculator closely corresponded to that obtained in Korean populations. Although the phenotype frequencies of several antigens differed significantly, the antigens with different frequencies compared to the previous data (by 2.1-3.7%) were B35, B62, and DR15. These discrepancies might be due to differences in population sampling or the HLA typing methods that were used. Since HLA-A, HLA-B, and HLA-DR antigen frequencies differ among various populations [11, 12], antigen frequencies in the CMC-CPRA calculator differed from those in the UNOS or ETRL programs. The most striking differences were the frequencies of HLA-A1, A3, A24, and A33 at the HLA-A locus.
Traditional PRA tests consist of a panel or pooled antigens, including either HLA class I or class II antigens, and represent the %PRA. In this study, we used the higher %PRA values based on either the class I or class II panel to compare with the %CPRA values. Although the PRA values from PRA-screen and PRA-ID tests and the CPRA values obtained from PRA-SA test varied (Fig. 2), the concordance rates for these were above 80% for the detection of broadly sensitized sera (PRA or CPRA levels greater than 50% or 80%). In a previous study, concordance was generally lower in the lower PRA groups due to an underestimation of sensitization using traditional PRA-ID testing [2]. In contrast, our study included a considerable number of cases that had higher PRA values than the CPRA in the lower CPRA groups. This may be due to less agreement for weaker antibodies, and additive or synergistic effects of multiple weak antibodies in the PRA-screen and PRA-ID assays. Because weak antibodies may not correlate with a positive flow cytometry crossmatch [13, 14], and appear to have little clinical importance [15-17], further studies will be necessary to establish the best strategy for predicting strong positive crossmatches [2]. It is also possible that the decreased agreement is due to the detection of anti-C or anti-DQ antibodies by the PRA-screen and PRA-ID tests. The HLA-C, HLA-DQ, and HLA-DP antigens are currently excluded from the CMC-CPRA calculation because adequate donor typing data to estimate their frequencies were not available. Since there were several reports indicating that donor-specific anti-C, anti-DQ, or anti-DP antibodies can induce antibody-mediated rejection [18-20], these antibodies should be included in the CPRA calculators in the future. In addition, allele-specific antibodies can be detected by PRA-SA tests, which can lead to a positive crossmatch when the target antigen is present in the donor. Therefore, it may be important to account for all HLA specificities in the CPRA calculator and to test donor HLA typing at the allele level, compatible with HLA antibody detection.
Although PRA and CPRA values had more than 80% agreement for the detection of broadly sensitized sera in this study, CPRA values would provide more consistent data for reporting sensitization than PRA values. Therefore, in KONOS, CPRA can be used to standardize sensitization measurements and can be used for standardized allocation of kidneys in an effort to compensate for the biological disadvantage of broadly sensitized patients. Moreover, high pretransplant PRA values (>50% or >80%) are known to be a poor prognostic marker in living donor renal allograft. Therefore, CPRA can also be used as a more accurate perioperative immunologic marker, and the adoption of virtual crossmatching using unacceptable antigens may increase the number of successful kidney transplantations in sensitized patients [21-23].
In conclusion, the application of unacceptable antigens and CPRA based on HLA phenotype frequencies in Koreans may be useful for the accurate and consistent measurement of sensitization and thus promoting efficient kidney allocation. Additional clinical studies are needed to confirm the benefit of CPRA calculations for organ allocation and successful transplantation.
Go to : 
Acknowledgement
This research was supported by Seoul St. Mary's Clinical Medicine Research Program during the year 2009 through the Catholic University of Korea.
The authors thank Dr. Sang-Hyun Hwang at the National Cancer Center for advice on the Hardy-Weinberg test.
Go to : 
References
1. Cecka JM, Kucheryavaya AY, Reinsmoen NL, Leffell MS. Calculated PRA: initial results show benefits for sensitized patients and a reduction in positive crossmatches. Am J Transplant. 2011; 11:719–724. PMID: 21114658.

2. Cecka JM. Calculated PRA (CPRA): the new measure of sensitization for transplant candidates. Am J Transplant. 2010; 10:26–29. PMID: 19958328.

3. Jung S, Oh EJ, Yang CW, Ahn WS, Kim Y, Park YJ, et al. Comparative evaluation of ELISA and Luminex panel reactive antibody assays for HLA alloantibody screening. Korean J Lab Med. 2009; 29:473–480. PMID: 19893358.

4. Bray RA, Gebel HM. Strategies for human leukocyte antigen antibody detection. Curr Opin Organ Transplant. 2009; 14:392–397. PMID: 19610172.

5. Howell WM, Carter V, Clark B. The HLA system: immunobiology, HLA typing, antibody screening and crossmatching techniques. J Clin Pathol. 2010; 63:387–390. PMID: 20418230.

6. Roh EY, Kim HS, Kim SM, Lim YM, Han BY, Park MH. HLA-A, -B, -DR allele frequencies and haplotypic associations in Koreans defined by generic-level DNA typing. Korean J Lab Med. 2003; 23:420–430.
7. Whang DH, Yang YS, Hong HK. Allele and haplotype frequencies of human leukocyte antigen-A, -B, and -DR loci in Koreans: DNA typing of 1,500 cord blood units. Korean J Lab Med. 2008; 28:465–474. PMID: 19127112.

8. Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995; 86:248–249.

9. Rousset F. Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008; 8:103–106. PMID: 21585727.

10. Zachary AA, Montgomery RA, Leffell MS. Defining unacceptable HLA antigens. Curr Opin Organ Transplant. 2008; 13:405–410. PMID: 18685337.

11. Lee KW, Kim YS. Serologic ambiguity and allelic frequency of the HLA-B40 family in the Korean population. Tissue Antigens. 1997; 49:383–388. PMID: 9151390.

12. Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens. 2005; 65:437–447. PMID: 15853898.

13. Kerman R, Lappin J, Kahan B, Katz S, McKissick E, Hosek K, et al. The crossmatch may still be the most clinically relevant histocompatibility test performed. Clin Transpl. 2007; 227–229. PMID: 18642454.
15. Phelan D, Mohanakumar T, Ramachandran S, Jendrisak MD. Living donor renal transplantation in the presence of donor-specific human leukocyte antigen antibody detected by solid-phase assay. Hum Immunol. 2009; 70:584–588. PMID: 19477211.

16. Aubert V, Venetz JP, Pantaleo G, Pascual M. Low levels of human leukocyte antigen donor-specific antibodies detected by solid phase assay before transplantation are frequently clinically irrelevant. Hum Immunol. 2009; 70:580–583. PMID: 19375474.

17. Ho EK, Vasilescu ER, Colovai AI, Stokes MB, Hallar M, Markowitz GS, et al. Sensitivity, specificity and clinical relevance of different cross-matching assays in deceased-donor renal transplantation. Transpl Immunol. 2008; 20:61–67. PMID: 18929659.

18. Bray RA, Murphey C, Schaub S. Calculated PRA: a process whose time has come or 'Déjà vu' all over again? Am J Transplant. 2011; 11:650–651. PMID: 21401870.

19. Vaidya S, Hilson B, Sheldon S, Cano P, Fernandez-Vina M. DP reactive antibody in a zero mismatch renal transplant pair. Hum Immunol. 2007; 68:947–949. PMID: 18191721.

20. Goral S, Prak EL, Kearns J, Bloom RD, Pierce E, Doyle A, et al. Preformed donor-directed anti-HLA-DP antibodies may be an impediment to successful kidney transplantation. Nephrol Dial Transplant. 2008; 23:390–392. PMID: 17956891.

21. Bray RA, Nolen JD, Larsen C, Pearson T, Newell KA, Kokko K, et al. Transplanting the highly sensitized patient: The emory algorithm. Am J Transplant. 2006; 6:2307–2315. PMID: 16939516.

22. Appel JZ 3rd, Hartwig MG, Cantu E 3rd, Palmer SM, Reinsmoen NL, Davis RD. Role of flow cytometry to define unacceptable HLA antigens in lung transplant recipients with HLA-specific antibodies. Transplantation. 2006; 81:1049–1057. PMID: 16612283.

23. Leffell MS, Cherikh WS, Land G, Zachary AA. Improved definition of human leukocyte antigen frequencies among minorities and applicability to estimates of transplant compatibility. Transplantation. 2007; 83:964–972. PMID: 17460569.

Go to : 




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download