Abstract
Background
In this study, we evaluated the analytical performance and clinical potential of a one-step multiplex real-time PCR assay for the simultaneous detection of 14 types of respiratory viruses using the AdvanSure RV real-time PCR Kit (LG Life Sciences, Korea).
Methods
Three hundred and twenty clinical specimens were tested with the AdvanSure RV real-time PCR Kit and conventional multiplex reverse transcription (RT)-PCR assay. The assay results were analyzed and the one-step AdvanSure RV real-time PCR Kit was compared with the conventional multiplex RT-PCR assay with respect to the sensitivity and specificity of the detection of respiratory viruses.
Results
The limit of detection (LOD) was 1.31 plaque-forming units (PFU)/mL for human rhinoviruses (hRVs), 4.93 PFU/mL for human coronavirus HCoV-229E/NL63, 2.67 PFU/mL for human coronavirus HCoV-OC43, 18.20 PFU/mL for parainfluenza virus 1 (PIV)-1, 24.57 PFU/mL for PIV-2, 1.73 PFU/mL for PIV-3, 1.79 PFU/mL for influenza virus group (Flu) A, 59.51 PFU/mL for FluB, 5.46 PFU/mL for human respiratory syncytial virus (hRSV)-A, 17.23 PFU/mL for hRSV-B, 9.99 PFU/mL for human adenovirus (ADVs). The cross-reactivity test for this assay against 23 types of non-respiratory viruses showed negative results for all viruses tested. The agreement between the one-step AdvanSure multiplex real-time PCR assay and the conventional multiplex RT-PCR assay was 98%.
Respiratory tract infection is an important common cause of hospitalization across all age groups [1, 2]. Infectious respiratory diseases in humans may be caused by several pathogens, including viruses and bacteria that produce very similar clinical symptoms and make diagnosis difficult [3, 4]. For this reason, an effective diagnostic assay for these pathogens in individual samples is urgently needed [5]. Furthermore, the 2003 severe acute respiratory syndrome (SARS) outbreaks and, more recently, the human and avian H5N1/H1N1 influenza virus cases underscore the importance of a rapid and accurate laboratory diagnostic method to characterize respiratory infections [6].
Clinical virology laboratories have used methods such as direct fluorescent-antibody assay (DFA) and virus culture to diagnose viral respiratory tract infections and identify respiratory viruses (RVs). These detection methods generally offer a rapid processing time but are labor intensive and require specific monoclonal antibodies. These methods are also limited by the availability of monoclonal antibodies for recently discovered viruses [7]. Moreover, these conventional and routine diagnostic assays have been shown to be inferior in sensitivity and specificity to nucleic acid-based assays including the PCR, which can be designed to screen for a wide range of pathogens [8, 9]. As a result, numerous recent studies have focused on the development and evaluation of multiplex PCR, reverse transcription (RT) PCR, or real-time PCR to diagnose both characterized and emerging RV infections with improved sensitivity [10-14].
Mono-specific PCR assays amplify each target in separate reactions and are, therefore, expensive and resource intensive. Multiplex PCR for clinical diagnosis uses a combination of several different primer pairs in the same amplification reaction simultaneously, depending on the targets present in the clinical sample [15]. Thus, multiplex PCR has been used increasingly for the diagnosis of infectious diseases, including the presence of one or more RV infections in respiratory tract specimens [16, 17]. Real-time PCR, with specific tracking of the product by fluorescent probes, improves assay specificity and significantly reduces hands-on time. In addition, real-time PCR offers the ability to perform multiplex amplification and detection. In some real-time PCR platforms, several different amplification products may be monitored in a single tube [18].
The one-step AdvanSure multiplex real-time PCR assay can simultaneously detect up to 14 different RVs. Specific targeted pathogen for this assay are 12 types of RNA viruses, i.e.,- several human coronaviruses (HCoV-229E, HCoV-NL63, HCoV-OC43), parainfluenza viruses 1, 2, 3 (PIV-1, PIV-2, PIV-3), influenza virus groups A and B (FluA and FluB), human respiratory syncytial viruses A and B (hRSV-A, hRSV-B), human rhinovirus (hRVs), human metapneumovirus (hMPV), and 2 types of DNA viruses - human adenoviruses (ADVs) and human bocavirus (HBoV), all in one step. In this study, we evaluated the analytical performance and clinical applicability of a recently developed one-step multiplex real-time PCR assay, the AdvanSure RV real-time PCR Kit (LG Life Sciences, Seoul, Korea), and compared the results to those of a conventional multiplex RT-PCR assay (Seeplex RV12 Detection Kit, Seegene, Seoul, Korea).
Between November 2008 and February 2010, 320 specimens were collected consecutively from patient samples submitted to the Dankook University Hospital for routine RV screening. The specimens, 310 nasopharyngeal aspirates (NPAs), 5 nasal swabs (NS), and 5 throat swabs (TS), were stored at -70℃ before nucleic acid extraction and multiplex PCR assays. All 320 specimens were analyzed by using the one-step multiplex real-time Advan Sure RV real-time PCR Kit (LG Life Science) and a conventional multiplex RT-PCR assay (Seeplex RV12 Detection Kit, Seegene) simultaneously.
The DNA was extracted from the clinical samples using the QIA-cube platform (Qiagen, Hilden, Germany). Extracted nucleic acids were then amplified and probed for RVs with the AdvanSure RV real-time PCR Kit according to the manufacturer's instructions. Briefly, 5 µL of extracted DNA was added in an AdvanSure RV real-time PCR reaction tube containing 5 µL of primer probe mixture and 10 µL of one-step RT-PCR premix. For the reverse transcription step, this mixture was incubated at 50℃ for 10 min. Denaturation followed at 95℃ for 30 sec, then 10 cycles of PCR (15 sec at 95℃, 30 sec at 53℃, and 30 sec at 60℃), and 30 additional cycles of PCR for the detection of fluorescence signals (15 sec at 95℃, 30 sec at 53℃, 30 sec at 60℃). As an internal control, the human RNase P (rnp) gene was quantified in all assayed samples.
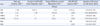
To determine the analytical sensitivity of the AdvanSure RV multiplex real-time PCR assay for each of the RVs, we measured the limit of detection (LOD) using 11 standard viruses as follows: HCoV-229E (KBPV-VR-9), HCoV-OC43 (KBPV-VR-8), PIV-1 (KBPV-VR-44), PIV-2 (KBPV-VR-45), PIV-3 (KBPV-VR-46), FluA (KBPV-VR-33), FluB (KBPV-VR-34), hRSV-A (KBPV-VR-41), hRVs (KBPV-VR-39), and ADVs (KBPV-VR-1) were purchased from the Korea Bank for Pathogenic Viruses (College of Medicine, Korea University, Seoul, Korea) and hRSV-B (ATCC-VR-1580) was purchased from ATCC. The sensitivity of the AdvanSure RV real-time PCR assay was determined for each of the viruses using 5-fold serial dilutions of virus suspensions in Microtest M4RT multi-microbe media (Remel, Lenexa, KS, USA) and referred to the concentration that was determined by the respective supplier (Table 1).
The sensitivity was quantified in terms of plaque-forming units (PFUs) per milliliter (PFU/mL). The PFU/mL result represents the number of infective virus particles within the sample on the basis of the assumption that each plaque formed is representative of 1 infective virus particle. The statistical analysis was conducted using the Minitab 16 statistical software (Minitab, State College, PA, USA). The LOD (95%) value of acquired data was compared to the previously reported LOD (95%) values of Food and Drug Administration (FDA)-approved commercial RV test kits as follows: Proflu plus (Gen-Probe Inc., San Diego, CA, USA) [19], Simplex Flu A/B & RSV (Focus Diagnostics, Cypress, CA, USA) [20], Verigene RV Nucleic Acid Test (Nanosphere, Northbrook, IL, USA) [21] and xTAG RVP FAST (Luminex, Austin, TX, USA) [22].
The cross-reactivity of the AdvanSure RV real-time PCR Kit was assessed using 23 types of non-RVs. Human T-lymphotropic virus (HTLV)-I and/or II-positive human plasma (DR1191), cytomegalovirus (CMV) IgG-positive human plasma (DV1030), Epstein-Barr virus (EBV) IgG-positive human plasma (DV1053), Herpes simplex virus (HSV) I and/or II-IgG positive human plasma (DV1081), Varicella zoster virus (VZV)-positive human plasma (DV1210) and HIV I and/or II-positive human plasma (DR1093) were supplied by Trina Bioreactives AG (Pembroke Pines, FL, USA). The CMV reference strain (KPBV-7) was obtained from the Korea Bank for Pathogenic Viruses. HCV-positive plasma (PHV205-16) was purchased from BBI Diagnostics (West Bridgewater, MA, USA). Enterobacter cloacae (CCARM-0298), Klebsiella pneumoniae (CCARM-10258), Pseudomonas aeruginosa (CCARM-0024), Staphylococcus aureus (CCARM-3A160), and Streptococcus pneumoniae (CCARM-4001) were obtained from Culture Collection of Antimicrobial Resistant Microbes (Seoul, Korea). Mycobacterium tuberculosis (M. tuberculosis) (00000-00021), M. avium (00136-41001), M. intracellulare (00000-00010), M. abscessus (00136-61010), M. ulcerans (00000-00029), M. scroflaceum (00000-00016), M. chelonae (00136-62001), M. kansasii (00136-20001), M. gordonae (00000-00008), and M. terrae (00000-00019) were purchased from Korea Mycobacterium Resource Center (Seoul, Korea). The RNA or DNA of supplied samples was extracted and assayed with the AdvanSure RV real-time PCR Kit adhering to the same procedures used for sample processing.
For the comparison of the sensitivity of the one-step AdvanSure RV real-time PCR and conventional multiplex RT-PCR assays, all 320 specimens which were used in the one-step multiplex real-time AdvanSure RV real-time PCR assay were tested by conventional multiplex RT-PCR assay using the Seeplex RV12 Detection Kit (Seegene) simultaneously.
Viral RNA was extracted from clinical samples with the QIAamp MinElute Virus Spin Kit (Qiagen) and used to synthesize cDNAs using the Revert Aid First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD, USA). Reverse transcriptions were performed for 90 min at 37℃ in a final reaction volume of 20 µL that consisted of 0.2 µg/µL random hexamer, 50 ng total RNA, 10 mM dNTP, 200 µg/µL reverse transcriptase, 20 µg/µL RNase inhibitor, and RT buffer. PCR amplification was performed using the Seeplex RV12 Detection Kit, according to the manufacturer's instructions, with a PTC200 PCR system (MJ Research, St. Bruno, Quebec, Canada). Briefly, PCR reactions were cycled in the following sequence 40 times: 94℃ for 30 sec, 60℃ for 90 sec, and 72℃ for 90 sec. The final cycle was followed by an extension step at 72℃ for 10 min to allow completion of any partial polymerization. The amplified PCR products were separated on a 2% agarose gel and stained with ethidium bromide. The type of RV was identified through comparison with reference band sizes provided by the manufacturer. As an internal control, plasmids containing amplicons of 719 bp were included during viral RNA/DNA extraction. The multiplex RT-PCR targeted genes for 11 types of RVs were as follows: HCoV-229E/NL63, HCoV-OC43, PIV-1, PIV-2, PIV-3, FluA, FluB, hRSV-A, hRSV-B, hRVs and ADVs.
Six samples that showed a discrepancy between the one-step AdvanSure assay and the conventional RT-PCR assay were further verified using nested real-time RT-PCR and sequencing using the primer sets included in the AdvanSure RV real-time PCR kit. The validation RT-PCR was performed using 10 µL of RT-PCR mix (Roche, Mannheim, Germany), 5 µL of outer primer set, and 5 µL of extracted sample DNA or RNA. After a reverse transcription step at 50℃ for 10 min and a denaturation step at 95℃ for 30 sec, 30 cycles of 3-step PCR (15 sec at 95℃, 30 sec at 53℃, and 30 sec at 60℃) were performed. The nested PCR was performed using 10 µL of HS Taq premix (Genetbio, Nonsan, Korea), 5 µL of inner primer and probe set, and 5 µL of 10-fold and 100-fold diluents of RT-PCR products. After denaturation for 10 min at 95℃, 30 cycles of 3-step PCR were performed (15 sec at 95℃, 30 sec at 53℃, and 30 sec at 60℃). During the nested PCR, the fluorescent signals were detected as part of real-time signal monitoring. For verification of the 6 samples with discrepancies, gene sequencing was performed by Geno-Tech (Daejeon, Korea). The PCR products, purified using the QIAquick PCR purification kit (Qiagen) were used as template DNA for bidirectional sequencing. The PCR for sequencing was performed using BigDye Terminator Cycle Sequencing and analyzed with an ABI 3100 Prism Automated DNA sequencer (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer's instructions.
The LODs (95%) of the one-step AdvanSure RV real-time PCR assay were determined for each of the following 11 viral targets from standard virus samples arranged in a dilution series from a high titer stock (Table 1). The acquired 95% LODs (PFU/mL) for the 11 standard virus were 4.93 for HCoV-229E/NL63, 2.67 for HCoV-OC43, 18.20 for PIV-1, 24.57 for PIV-2, 1.73 for PIV-3, 1.79 for FluA, 59.51 for FluB, 5.46 for hRSV-A, 17.23 for hRSV-B, 9.99 for ADVs and 1.31 for hRVs (Table 2). As shown in Table 2, detection of hRVs was the most sensitive among the 11 targeted standard viruses and the LODs of FluB, PIV-1, PIV-2, and hRSV-B showed decreased sensitivity compared to the LODs of the other viruses.
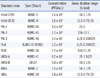
The assay sensitivity was also compared with that of several previously reported US FDA-approved commercial RV test kits (Table 3). The LOD assay results showed that our AdvanSure real-time PCR assay has similar or higher sensitivity than commercially available RV assays in the viruses tested, except for FluB and hRSV-B. FluB and hRSV-B exhibited higher LOD values in our assay, compared to other compared commercial RV test kits.
To evaluate the cross-reactivity and detection specificity, 23 different non-RV reference strains were tested using the same assay procedure used for the clinical samples for RV detection with the AdvanSure RV real-time PCR detection Kit. All assay results were negative and no non-specific positive reaction was observed (Table 4).
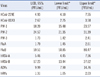
The AdvanSure RV multiplex real-time PCR assay and conventional multiplex RT-PCR assay were performed on 320 clinical samples simultaneously. Among the 320 samples, 143 positive samples were detected by the AdvanSure RV multiplex real-time PCR assay and 141 positive samples detected by the conventional multiplex RT-PCR. Most of the viruses detected by conventional methods were detected by the AdvanSure multiplex assay. Though both assays showed an outstanding ability to detect RVs, 4 samples that were identified as positive samples by the AdvanSure assay, including 2 FluA-positive, 1 HCoV-NL63-positive, and 1 hRVs-positive, were identified as negative samples by the conventional RT-PCR assays. On the other hand, 2 clinical samples that were negative in the AdvanSure multiplex real-time PCR assay were identified as positive by the conventional multiplex RT-PCR assay (Table 5).
For further analysis of the 6 samples that yielded discrepant results with the AdvanSure assay and the conventional multiplex RT-PCR assay, uniplex nested real-time RT-PCR and gene sequencing were performed. The results of the nested real-time RT-PCR assay and gene sequencing revealed that all samples that were identified as positive from either of the assays were indeed accurate positive results, irrespective of their classification as negative by either assay (Table 5).
A variety of clinical diagnostic assays utilizing different methods and targets for RV assessment have been reported, but cell culture is still considered the "gold standard". However, this method has drawn criticism for difficulties such as maintaining cell cultures, instability of cultured cells, and a long turnaround cycle, from 1 to as many as 14 days [23-26]. The prolonged turnaround time of this diagnostic method for confirming RVs in clinical samples is an obstacle to rapid initiation of antiviral therapy, isolation of infected patients, and/or cessation of the unnecessary use of antibiotics [27]. The rapid diagnosis of viral infections has depended mainly on viral antigen detection; however, the sensitivities of these assays vary (50-90%), depending on the assay method and the virus of interest [28]. For these reasons, rapid, easy-to-perform, sensitive, specific, and cost-effective diagnostic techniques are increasingly needed in the clinical microbiology laboratory.
Multiplex real-time RT-PCR is a recognized technique offering a faster and effective technology for the rapid detection of viruses than the conventional diagnostic methods of virus culture, antigen tests, or direct immunofluorescence [29]. In addition, multiplex RT-PCR assays that are able to target several types of respiratory pathogens have been reported previously [30, 31]. In this study, we evaluated the performance of the one-step AdvanSure multiplex real-time PCR assay for common respiratory viral pathogens, which include HCoV-229E/NL63, HCoV-OC43, PIV-1, PIV-2, PIV-3, FluA, FluB, hRSV-A, hRSV-B, ADVs, and hRVs and compared it to that of a conventional multiplex RT-PCR assay. The one-step AdvanSure assay provides a faster turnaround time because it avoids additional nested amplification or hybridization steps needed for identification of viral products. Moreover, the AdvanSure assay minimizes the possibility of contamination as it eliminates the need for additional post-PCR processing of the samples and the 11 viral targets could be detected simultaneously.
The analytical sensitivity of the AdvanSure assay demonstrated its clinical potential, in particular for diagnosis of HCoV-229E, HCoV-NL63, PIV-3, FluA, hRSV-A, ADVs, and hRVs (1-10 PFU/mL). These LOD levels for RVs are similar to values previously reported by other investigators [30, 32]. The LOD levels for each of the viral targets have also been reported for FDA-approved RV assay kits [19-22]. Although the AdvanSure assay had higher LOD values for FluB and hRVs than those of the FDA-approved RV assay kits, it had lower LOD values for ADVs. For other viral targets, the LODs from the AdvanSure assay proved to be comparable to those associated with the FDA-approved RV assay kits. In general, the multiplex RT-PCR assay for the detection of RVs supports the proper primer against the RVs and their mutations and the sensitivity of the RT-PCR assays depend on the degree of sequence identity/similarity between the primers and the genomes of the tested viruses [33, 34]. In this study, the higher LODs obtained for FluB and hRVs determinations likely reflect unsuitable or sub-optimal primer sequences for these viruses in the AdvanSure assay kit against the tested standard viruses.
Our evaluation of specificity or cross-reactivity in the AdvanSure assay against 23 reference strains indicated that none of the samples gave a false positive reaction. These outstanding results obtained from our trial of the AdvanSure assay clearly demonstrate its clinical potential compared with the specificities recently reported for alternative detection methods [33-36].
To determine the clinical performance of the one-step AdvanSure multiplex real-time PCR assay for the RVs, we compared the results of the AdvanSure assay to those of a conventional multiplex RT-PCR assay by using 320 collected clinical samples. The sensitivity of the conventional multiplex RT-PCR was 44.06% (141/320), while the sensitivity of the AdvanSure assay was 44.68% (143/320). The main advantage of the AdvanSure assay is the ability to identify 2 additional RVs not detected by the conventional RT-PCR assay, including hMPVs and HBoV. Although, the 2 kinds of multiplex RT-PCR assays showed equivalent positive rates overall, 6 samples that had different results with the 2 assays were identified. Two samples that were positive for FluA, 1 for HCoV-NL63, and 1 for hRVs by the AdvanSure assay were identified as negative by the conventional RT-PCR assay. Two samples that were positive for PIV-2 by the conventional multiplex RT-PCR assay were identified as negative by the AdvanSure assay. For resolving the discrepancies between the 2 assays for these 6 samples, nested real-time PCR and gene sequencing were performed to confirm that the results were indeed true positive and not false-positive. The results of nested RT-PCR and gene sequencing indicated that the samples classified as positive by either assay were indeed positive, and that both assays had, therefore, yielded false-negative results for these samples. This discrepancy may result from the limitations of multiplex PCR assays. One of the major limitations is the assay principle of multiplex PCRs, which could produce false results if a primer region has nucleotide variations and is unable to detect new types or strains of a virus. This is the reason that direct antigen tests or virus cultures cannot be completely replaced by multiplex PCR assays [33]. Another limitation is the false positive result as a consequence of exposure to amplified PCR products during a conventional 2-step RT-PCR [28]. Although the conventional multiplex RT-PCR kit used in this study featured a dual priming oligonucleotide system, it still yielded higher false-positive results than the one-step AdvanSure assay, and these findings agree with those of a previous publication [37].
In conclusion, the AdvanSure RV real-time PCR assay demonstrated excellent overall sensitivity and specificity. Furthermore, the results of this assay may be obtained within 4 h, requiring minimal technician time. It offers clinical laboratories a valuable option for detection of RVs potentially improving clinical management with earlier diagnosis and treatment.
Figures and Tables
Table 3
List of LOD (95%) for respiratory viruses in the one-step AdvanSure real-time PCR assay and FDA-approved commercial RV test kits
Acknowledgement
The present research was conducted by the research fund of Dankook University in 2010.
References
1. Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998. 36:2990–2995.

2. Welliver RC. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J Pediatr. 2003. 143:5 Suppl. S112–S117.

3. Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004. 29:179–188.

4. McDonough EA, Barrozo CP, Russell KL, Metzgar D. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol Cell Probes. 2005. 19:314–322.
5. Adcock PM, Stout GG, Hauck MA, Marshall GS. Effect of rapid viral diagnosis on the management of children hospitalized with lower respiratory tract infection. Pediatr Infect Dis J. 1997. 16:842–846.

6. Lam WY, Yeung AC, Tang JW, Ip M, Chan EW, Hui M, et al. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007. 45:3631–3640.

7. Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007. 45:2965–2970.

8. Casiano-Colon AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol. 2003. 28:169–174.
9. Falsey AR, Criddle MC, Walsh EE. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J Clin Virol. 2006. 35:46–50.

10. Boivin G, Côte S, Déry P, De Serres G, Bergeron MG. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J Clin Microbiol. 2004. 42:45–51.

11. Strålin K, Bäckman A, Holmberg H, Fredlund H, Olcén P. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS. 2005. 113:99–111.
12. Vabret A, Mouthon F, Mourez T, Gouarin S, Petitjean J, Freymuth J. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J Virol Methods. 2001. 97:59–66.

13. Lim G, Park TS, Suh JT, Lee HJ. Comparison of R-mix virus culture and multiplex reverse transcriptase-PCR for the rapid detection on respiratory viruses. Korean J Lab Med. 2010. 30:289–294.
14. Kim JS, Lim CS, Kim YK, Lee KN, Lee CK. Human bocavirus in patients with respiratory tract infection. Korean J Lab Med. 2011. 31:179–184.

15. Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004. 42:1564–1569.

16. Coiras MT, Aguilar JC, García ML, Casas I, Pérez-Breña P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol. 2004. 72:484–495.

17. Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean-Lecherbonnier J, et al. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005. 126:53–63.

18. Hu A, Colella M, Tam JS, Rappaport R, Cheng SM. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real-time PCR. J Clin Microbiol. 2003. 41:149–154.

19. FDA. Substantial equivalence determination decision summary assay only template. Updated on 2007. http://www.accessdata.fda.gov/cdrh_docs/reviews/K073029.pdf.
20. FDA. 510(k) Summary. Simplexa™ Flu A/B & RSV. Updated on Nov 2010. http://www.accessdata.fda.gov/cdrh_docs/pdf10/K102170.pdf.
21. FDA. Summary of 510(k) Safety and Effectiveness. Updated on May 2009. http://www.accessdata.fda.gov/cdrh_docs/pdf8/K083088.pdf.
22. FDA. 510(k) Substantial Equivalence determination decision summary. Updated on July 2011. http://www.accessdata.fda.gov/cdrh_docs/pdf10/K103776.pdf.
23. Woo PC, Chiu SS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997. 35:1579–1581.

24. Leland DS, Emanuel D. Laboratory diagnosis of viral infections of the lung. Semin Respir Infect. 1995. 10:189–198.
25. Ghosh S, Champlin RE, Englund J, Giralt SA, Rolstone K, Raad I, et al. Respiratory syncytial virus upper respiratory tract illnesses in adult blood and marrow transplant recipients: combination therapy with aerosolized ribavirin and intravenous immunoglobulin. Bone Marrow Transplant. 2000. 25:751–755.

26. Hayden FG, Treanor JJ, Betts RF, Lobo M, Esinhart JD, Hussey EK. Safety and efficacy of the neuraminidase inhibitor GG167 in experimental human influenza. JAMA. 1996. 275:295–299.

27. Bruijnesteijn van Coppenraet LE, Swanink CM, van Zwet AA, Nijhuis RH, Schirm J, Wallinga JA, et al. Comparison of two commercial molecular assays for simultaneous detection of respiratory viruses in clinical samples using two automatic electrophoresis detection systems. J Virol Methods. 2010. 169:188–192.
28. Klaschik S, Lehmann LE, Raadts A, Hoeft A, Stuber F. Comparison of different decontamination methods for reagents to detect low concentrations of bacterial 16S DNA by real-time-PCR. Mol Biotechnol. 2002. 22:231–242.

29. Gunson RN, Collins TC, Carman WF. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J Clin Virol. 2005. 33:341–344.

30. Bolotin S, Lombos E, Yeung R, Eshaghi A, Blair J, Drews SJ. Verification of the Combimatrix influenza detection assay for the detection of influenza A subtype during the 2007-2008 influenza season in Toronto, Canada. Virol J. 2009. 6:37.

31. Kehl SC, Henrickson KJ, Hua W, Fan J. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J Clin Microbiol. 2001. 39:1696–1701.

32. Pehler-Harrington K, Khanna M, Waters CR, Henrickson KJ. Rapid detection and identification of human adenovirus species by adenoplex, a multiplex PCR-enzyme hybridization assay. J Clin Microbiol. 2004. 42:4072–4076.

33. Lee JH, Chun JK, Kim DS, Park Y, Choi JR, Kim HS. Identification of adenovirus, influenza virus, parainfluenza virus, and respiratory syncytial virus by two kinds of multiplex polymerase chain reaction (PCR) and a shell vial culture in pediatric patients with viral pneumonia. Yonsei Med J. 2010. 51:761–767.

34. Rand KH, Rampersaud H, Houck HJ. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011. 49:2449–2453.

35. Zaghloul MZ. Human bocavirus (HBoV) in children with respiratory tract infection by enzyme linked immunosorbent assay (ELISA) and qualitative polymerase chain reaction (PCR). Virol J. 2011. 8:239–243.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download