Abstract
We report a recent case in which ciprofloxacin-resistant Shigella flexneri was isolated from a 23-yr-old female patient with a history of travel to India. Prior to her admission to our internal medicine department, she experienced symptoms of high fever and generalized weakness from continuous watery diarrhea that developed midway during the trip. S. flexneri was isolated from the stool culture. Despite initial treatment with ciprofloxacin, the stool cultures continued to show S. flexneri growth. In the susceptibility test for antibiotics of the quinolone family, the isolate showed resistance to ciprofloxacin (minimum inhibitory concentration [MIC], 8 µg/mL), norfloxacin (MIC, 32 µg/mL), ofloxacin (MIC, 8 µg/mL), nalidixic acid (MIC, 256 µg/mL), and intermediate resistance to levofloxacin (MIC, 4 µg/mL). In molecular studies for quinolone resistance related genes, plasmid borne-quinolone resistance genes such as qnrA, qnrB, qnrS, aac(6')-Ib-cr, qepA, and oqxAB were not detected. Two mutations were observed in gyrA (248C→T, 259G→A) and 1 mutation in parC (239G→T). The molecular characteristics of the isolated S. flexneri showed that the isolate was more similar to the strains isolated from the dysentery outbreak in India than those isolated from Korea.
Shigella flexneri is an endemic organism in most developing countries and causes severe illnesses; the mortality rate of S. flexneri infections is higher than those of infections with other Shigella species [1]. Patients are often treated with antimicrobial agents to reduce the duration of the illness, and the treatment is known to reduce the period of Shigella excretion [2]. Quinolones, especially ciprofloxacin, are commonly used for the treatment of shigellosis. However, in recent years, some Shigella strains have rapidly developed resistance to antimicrobial agents [3]. In India, one of the endemic areas for S. flexneri, this organism is one of the predominant species isolated from cultures and shows 45.6% resistance to ciprofloxacin [4]. In 8 Asian countries, 12% of the S. flexneri isolates obtained from 2001 to 2004 were documented to show resistance to ciprofloxacin [5]. In Korea, the National Institute of Health reported 2 quinolone-resistant strains of S. flexneri among 5,938 Shigella spp. in 2008 [6]. Here, we present a recent case in which quinolone-resistant S. flexneri was isolated from a patient with a history of travel to India.
A 23-yr-old woman had a history of high fever with a chief complaint of general weakness induced by repeated watery diarrhea. She had been in India for 10 days prior to her admission to our hospital. She experienced these symptoms midway through her trip and subsequently received fluid treatment. When she visited our hospital, diarrhea and other symptoms were absent; however, she was examined for further evaluation. Her physical examination showed that the heart rate was 64 beats/min, respiratory rate was 20 breaths/min, and the blood pressure was 110/60 mmHg. Results of laboratory investigations were as follows: hemoglobin, 11.9 g/dL; white blood cell count, 5.72×109 cells/L (37% segmented neutrophils, 48% lymphocytes, 5% monocytes, 8% eosinophils, and 2% atypical lymphocytes); and serum sodium concentration, 141 mEq/L. Her stool occult blood cultures yielded no growth.
S. flexneri were abundant in the first stool culture specimen. The micro-broth dilution assay with MicroScan WalkAway 96 SI (SIEMENS, Sacramento, CA, USA) showed that the isolate was resistant to ampicillin, ampicillin/sulbactam, ciprofloxacin, and trimethoprim-sulphamethoxazole, but was susceptible to amikacin, cefepime, cefotetan, cefoxitin, cephalothin, gentamicin, imipenem, and piperacillin/tazobactam. The minimum inhibitory concentration (MIC) of ciprofloxacin was 8 µg/mL. Initially, the patient received ciproctan (ciprofloxacin) 500 mg three times a day over 5 days. However, S. flexneri were still abundant in the follow-up stool culture, and the antibiotic-susceptibility test results also showed the same pattern from the previous results. After introduction of ceftriaxone in the treatment regimen for 5 days, S. flexneri was not isolated from the stool specimen.
Quinolone susceptibility of the isolates was tested in accordance with the CLSI guidelines (Table 1). The MICs of quinolones were tested on Mueller-Hinton broth (Becton Dickinson, Baltimore, MD, USA) using the micro-broth dilution method with a 96-well cell culture plate (SPL Lifescience, Pocheon, Korea) [7]. E. coli ATCC 25922 was used as the control strain. The quinolone-type antibiotics tested included ciprofloxacin (Sigma Chemical Co., St. Louis, MO, USA), norfloxacin (Sigma Chemical Co.), ofloxacin (Sigma Chemical Co.), sparfloxacin (Sigma Chemical Co.), nalidixic acid (Sigma Chemical Co.), and levofloxacin (Jeil Pharmaceutical Co., Seoul, Korea).
Further molecular studies, which included inspection of mutations in the quinolone target genes and the genes responsible for the expression of the efflux pump (Table 2) were performed to determine the mechanism of resistance to quinolone. Each target gene was amplified using the PCR method, while the sequencing of gyrA [8], gyrB [9], parC [8], parE [9], qnrA [6], qnrB [6], qnrS [6], oqxA [10], oqxB [10], and aac(6')-Ib-cr [6] genes was conducted by SCL (Seoul Clinical Laboratories), with a 3130XL DNA genetic analyzer (Applied Biosystems, Foster City, CA, USA). Plasmid-borne quinolone resistance genes such as qnrA, qnrB, qnrS, aac(6')-Ib-cr, qepA, and oqxAB were absent. Two mutations (248C→T, 259G→A) were found in gyrA and 1 mutation was found in parC (239G→T).
A laboratory test was conducted to study the expression of mdfA, tolC, and ydhE, which play a role in the encoding of the efflux pump genes, by the Korea Center for Disease Control and Prevention (KCDC) under the National Institute of Health (NIH) (Fig. 1). Isolates were cultured in 5 mL of Mueller-Hinton Broth (Becton Dickinson, Franklin Lakes, NJ, USA) with or without 5 µg/mL of ciprofloxacin at 37℃ for 30 min [6]. Total RNA was isolated using the RNeasy Mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's protocol. Expression of each gene was determined by real-time PCR quantification, which was based on the relative expression levels of mdfA, tolC, and ydhE when compared with the expression levels of a reference gene for 16S rRNA. The sample was quantified by PCR in a final reaction volume of 20 µL, containing 2.5 µL of cDNA with TaqMan One-Step RT-PCR master mix (Applied Biosystems) [6]. Quantification was performed on the basis of the threshold cycle (CT) value, which was determined using the LC480 software system (Roche Diagnostics, Mannheim, Germany). Relative gene expression levels were calculated as 2-ΔΔCT, where ΔΔCT =ΔCT (sample) -ΔCT (control), and ΔCT is the CT of the target gene subtracted from the CT of the housekeeping gene [11]. When ciprofloxacin was included in the medium, the expression levels of tolC of the S. flexneri isolate increased to 3.7 times than the expression levels in the control isolate. S. flexneri isolated from NIH was used as the control isolate, which had only one mutation in gyrA (248C→T) with susceptibility to ciprofloxacin (MIC, 0.25 µg/mL).
Since 1998, cases of shigellosis caused by Shigella sonnei and S. flexneri have been steadily increasing in Korea [6], although cases of quinolone-resistant S. flexneri infections are still relatively rare. Several mechanisms are responsible for the development of quinolone resistance in gram-negative organisms. One of the prominent mechanisms is the mutation of the genes related to quinolone resistance. The target regions where mutations occur in the organism have been identified as topoisomerase IV (parC and parE) and DNA gyrase (gyrA and gyrB) [12]. Dynamic efflux of quinolone has also been observed, and effluxmodulated resistance has been reported in numerous pathogenic gram-negative bacteria [13]. Another mechanism depends on plasmid-mediated quinolone resistance (PMQR) determinants such as qnr, aac(6')-Ib-cr, and qepA [14].
In the 2 previously reported quinolone-resistant strains of S. flexneri in Korea [6], one strain was documented to contain 3 substitutions: 1 in gyrA (Ser83→Leu) and 2 in parC (Ser80→Ile and Arg91→Gln). The other strain had 4 substitutions: 2 in gyrA (Ser83→Leu and Asp87→Gly) and 2 in parC (Ser80→Ile and Arg91→Gln). Both the strains had the PMQR gene (qnrS) in common, and there was an increase in the expression levels of the efflux pump-encoding genes (mdfA, tolC, and ydhE), which were better expressed in the presence of ciprofloxacin [6] (Table 3). In the present case, S. flexneri showed the mutation of the genes related to quinolone resistance. Two mutations were documented in gyrA; 1 for nucleotide position 248C→T (Ser83→Leu) and the other for position 259G→A (Asp87→Asn). The change of amino acid codon 87 from aspartic acid to asparagine shows new amino acid substitution that has not been reported in the isolated strains in Korea. In parC gene, a mutation for nucleotide position 239G→T (Ser80→Ile) was also found. PMQR determinants were not found in the molecular analysis.
According to the reports on the dysentery outbreaks in India in 2008, all isolated Shigella spp. were susceptible to ceftriaxone, a few of the quinolone-resistant strains had the aac(6')-Ib-cr gene, and none of the documented strains had qnrA, B, and S genes [15]. In addition, quinolone-resistant S. flexneri strains had 2 documented mutations in gyrA and 1 mutation in parC, which is similar to the corresponding findings in our case [15]. When the strains were treated with CCCP (carbonyl cyamide-m-chlorophenyldrazone), norfloxacin or ciprofloxacin accumulation was noted in the cells. This suggests that efflux pumps are one of the factors responsible for the development of quinolone resistance. In our case, involvement of the efflux pump in quinolone resistance was evaluated by real-time PCR analysis. The expression levels of the efflux pump-encoding gene, tolC, were higher in the presence of ciprofloxacin than in its absence.
We report a case of shigellosis caused by quinolone-resistant S. flexneri , isolated from a patient who traveled to India. Although the mutation points in the gyrA and parC genes were identical to the mutation points in the strains isolated in Korea, the molecular characteristics of the isolate in our case were more similar to those of the quinolone-resistant strains isolated in India because amino acid substitutions in the strains are different and there were no PMQR genes.
Figures and Tables
Fig. 1
Expression of the efflux pump genes in response to ciprofloxacin with reference to the expression levels in the control strain. Expression of tolC, mdfA, and ydhE was measured by real-time PCR in the absence and presence of ciprofloxacin. Each value represents the average of 5 culture replicates, each of which was evaluated twice. Relative gene expression levels were calculated as 2-ΔΔCT, where ΔΔCT=ΔCT (sample) - ΔCT (control) [11].
CIP-: absence of ciprofloxacin; CIP+: presence of ciprofloxacin.
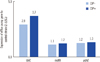
Table 3
Comparison between previously reported strains (strain number 1 and 2) and strain of present case (strain number 3)

*Previously reported S. flexneri strains in Korea [6].
References
1. Haukka K, Siitonen A. Emerging resistance to newer antimicrobial agents among Shigella isolated from Finnish foreign travellers. Epidemiol Infect. 2008. 136:476–482.
2. Salam MA, Bennish ML. Antimicrobial therapy for shigellosis. Rev Infect Dis. 1991. 13:Suppl 4. S332–S341.

3. Niyogi SK. Shigellosis. J Microbiol. 2005. 43:133–143.
4. Taneja N. Changing epidemiology of shigellosis and emergence of ciprofloxacin-resistant Shigellae in India. J Clin Microbiol. 2007. 45:678–679.
5. Kuo CY, Su LH, Perera J, Carlos C, Tan BH, Kumarasinghe G, et al. Antimicrobial susceptibility of Shigella isolates in eight Asian countries, 2001-2004. J Microbiol Immunol Infect. 2008. 41:107–111.
6. Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin Microbiol Infect. 2008. 14:760–765.
7. Clinical and Laboratory Standards Institute. Eighteenth Informational supplement, M100-S16. Performance standards for antimicrobial susceptibility testing. 2008. Wanye, PA: Clinical and Laboratory Standards Institute.
8. Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009. 53:235–241.
9. Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003. 47:3222–3232.
10. Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother. 2009. 53:3582–3584.
11. Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001. 25:386–401.

12. Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997. 61:377–392.

13. Poole K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin Microbiol Infect. 2004. 10:12–26.

14. Jacoby G, Cattoir V, Hooper D, Martínez-Martínez L, Nordmann P, Pascual A, et al. qnr Gene nomenclature. Antimicrob Agents Chemother. 2008. 52:2297–2299.
15. Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008. 57:856–863.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download