Abstract
In recent years, there have been increasing reports of KPC-producing Klebsiella pneumoniae in Korea. The modified Hodge test can be used as a phenotypic screening test for class A carbapenamase (CAC)-producing clinical isolates; however, it does not distinguish between carbapenemase types. The confirmation of type of CAC is important to ensure optimal therapy and to prevent transmission. This study applied a novel multiplex PCR assay to detect and differentiate CAC genes in a single reaction. Four primer pairs were designed to amplify fragments encoding 4 CAC families (SME, IMI/NMC-A, KPC, and GES). The multiplex PCR detected all genes tested for 4 CAC families that could be differentiated by fragment size according to gene type. This multiplex PCR offers a simple and useful approach for detecting and distinguishing CAC genes in carbapenem-resistant strains that are metallo-β-lactamase nonproducers.
Carbapenems are important antibiotics for the treatment of infections caused by multidrug-resistant gram-negative bacilli [1, 2]; however, carbapenem-resistance is increasing, causing infections that are difficult to treat [2-4]. Bacterial production of carbapenemases is one of the most important mechanisms of carbapenem resistance [1]. There are 3 molecular classes of carbapenemases: A (penicillinases); B (metallo-β-lactamases, MBLs); and D (oxacillinases). The class A carbapenemases (CACs) include the SME, IMI/NMC-A, SFC, BIC, KPC, and some type of GES family proteins. The genes for the SME, IMI/NMC-A (except IMI-2), SFC, and BIC enzymes are chromosomal, and the genes for KPC and GES are carried on plasmids [1]. KPC producers have caused severe treatment problems in hospitals around New York and have also been reported in Europe, South America, and China [5-8]. In Korea, KPC-producing Klebsiella pneumoniae have rarely been detected. However, several cases were reported in 2010 and 2011 (Interscience Conference on Antimicrobial Agents and Chemotherapy 2011 poster c2-652, unpublished observation) [9, 10].
The modified Hodge test (MHT) can be used as a phenotypic confirmatory test for suspected carbapenemase production in Enterobacteriaceae [11]. However, it is reported that the MHT shows approximately 25% false positive results among carbapenemase nonproducers, mainly AmpC hyperproducers and strains harboring CTX-M [12]. Moreover, it does not distinguish between carbapenemase types or CAC types.
The major concern from the therapeutic and epidemiologic perspective is with transmissible and not chromosomal carbapenemases [1, 2], and this information cannot be acquired by the phenotypic methods. Confirmation of the CAC type is important to ensure optimal therapy and to prevent transmission [3]. In this study, we developed a multiplex PCR assay to detect and differentiate multiple CAC genes in a single reaction.
Eleven CAC producers (1 SME-producing Serratia marcescens, 2 IMI/NMC-A-producing Enterobacter cloacae, 2 KPC-producing Enterobacteriaceae, and 6 GES-producing Klebsiella pneumoniae); 7 MBL producers (3 VIM-producing Pseudomonas aeruginosa, 2 IMP-producing P. aeruginosa, 1 IMP-producing Acinetobacter baumannii, and 1 SIM-producing A. baumannii); and 5 non-carbapenemase-producing Enterobacteriaceae were studied (Table 1).
The bacterial cells were lysed by heating at 95℃ for 10 min, and cellular debris was removed by centrifugation at 13,000 rpm for 5 min. The supernatant was used as the source of amplification templates. PCR was performed with a final volume of 20 µL in 0.2 mL thin-walled tubes (Accupower™ HotStart PCR PreMix; Bioneer, Daejeon, Korea).
We designed 4 primer pairs for 4 CAC families (SME, IMI/NMC-A, KPC, and GES). The genes encoding IMI and NMC-A type CACs are similar to each other and could not be differentiated by conventional PCR. Therefore, a single primer pair for the detection of these 2 CAC families was designed. The SFC-1 and the BIC-1 enzymes have been found in environmental isolates, and the corresponding genes are chromosomally encoded [13, 14]. For this reason, we did not design pairs of primers for these genes. The primers used in this study were GES primers for blaGES1-9 and blaGES11-20 (GES-F: 5'-GCTTCATTCACGCACTATT-3'; GES-MR: 5'-CGATGCTAGAAACCGCTC-3'; product size: 323 bp), IMI/NMC-A primers for blaIMI1-3 and blaNMC-A (IMI(NMC)-F1: 5'-TGCGGTCGATTGGAGATAAA-3'; IMI(NMC)-R1: 5'-CGATTCTTGAAGCTTCTGCG-3'; product size: 399 bp), SME primers for blaSME1-3 (SME-F1: 5'-ACTTTGATGGGAGGATTGGC-3'; SME-R1: 5'-ACGAATTCGAGCATCACCAG-3'; product size: 551 bp), and KPC primers for blaKPC2-13 (KPCF2: 5'-GTATCGCCGTCTAGTTCTGC-3'; KPCFR: 5'-GGTCGTGTTTCCCTTTAGCC-3'; product size 638 bp). The PCR program consisted of an initial denaturation step at 94℃ for 5 min, followed by 25 cycles of DNA denaturation at 94℃ for 30 sec, primer annealing at 50℃ for 30 sec, and primer extension at 72℃ for 1 min. After the last cycle, a final extension step at 72℃ for 7 min was added.
The GenBank nucleotide sequence accession numbers for the sequences studied here were as follows: GES-1 (AF156486); GES-2 (AF326355); GES-3 (AB113580); GES-4 (AB116260); GES-5 (AY494717); GES-6 (AY494718); GES-7 (IBC-1, AF208529); GES-8 (IBC-2, AF329699); GES-9 (AY920928); GES-11 (FJ854362); GES-12 (FN554543); GES-13 (GU169702); GES-14 (GU207844); GES-15 (GU208678); GES-16 (HM173356); GES-17 (HQ874631); GES-18 (JQ028729); GES-19 (JN596280); GES-20 (JN596280); IMI-1 (U50278); IMI-2 (DQ173429); IMI-3 (GU015024); NMC-A (Z21956); SME-1 (Z28968); SME-2 (AF275256); SME-3 (AY584237); KPC-2 (AY034847); KPC-3 (AF395881); KPC-4 (AY700571); KPC-5 (EU400222); KPC-6 (EU555534); KPC-7 (EU729727); KPC-8 (FJ234412); KPC-9 (FJ624872); KPC-10 (GQ140348); KPC-11 (HM066995); KPC-12 (HQ342889); and KPC-13 (HQ342890).
The CAC families could be differentiated into 4 groups, SME, IMI/NMC-A, KPC, and GES, by the PCR product size (Fig. 1). None of the non-CAC producers included in this study produced PCR product bands. Not all genotypes of CAC were tested: only SME-1, IMI-1, NMC-A, KPC-2, KPC-3, and GES-5-type enzyme-producing strains were included in this study. The primers for the genotypic detection of SME, KPC and GES enzymes were exactly complementary to the corresponding GenBank sequences, but the primers for IMI/NMC-A were not complementary at 1 base each in the forward and reverse sequences of IMI-3 and NMC-A. Therefore, it is somewhat uncertain whether this multiplex PCR assay would be able to detect all of the genotypes of CACs described above.
In summary, this multiplex PCR method appears to be a simple and useful approach for detecting and distinguishing CAC genes in MBL-negative carbapenem-resistant strains. Therefore, this method should be helpful for characterization of CACs and prevention of the spread of pathogens producing these enzymes.
Figures and Tables
Fig. 1
Results of multiplex PCR for class A carbapenemase (CAC)-producing strains. Multiplex PCR products were separated on a 2% agarose gel. Lanes 1 and 14 show the 100-bp DNA ladder; lane 2, the PCR product of the negative control (distilled water); lanes 3 and 4, KPC-type enzyme-producing strains; lane 5, SME-type; lanes 6 and 7, NMC-A and IMI-type, respectively; lanes 8-13, GES-type. The amplified product from each PCR is indicated on the right, and the size of the marker in base pairs is shown on the left.
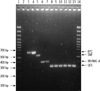
Table 1
Bacterial strains used for class A carbapenemase multiplex PCR

*ESBLs were detected by CLSI phenotypic confirmatory tests and type specific PCR; †CRAB, Center for Research in Anti-Infectives and Biotechnology, Department of Medical Microbiology and Immunology, School of Medicine, Creighton University, Omaha, Nebraska; ‡EBC is a group of AmpC β-lactamase originated from E. cloacae.
Abbreviation: ESBL, extended-spectrum β-lactamase.
Acknowledgement
We thank Dr. Kenneth S. Thomson (Center for Research in Anti-Infectives and Biotechnology, Department of Medical Microbiology and Immunology, School of Medicine, Creighton University, Omaha, Nebraska) for providing class A carbapenemase-producing isolates.
References
1. Patel G, Bonomo RA. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther. 2011. 9:555–570.

2. Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011. 53:60–67.
3. Miriagou V, Cornaglia G, Edelstein M, Galani I, Giske CG, Gniadkowski M, et al. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin Microbiol Infect. 2010. 16:112–122.

4. Cornaglia G, Rossolini GM. The emerging threat of acquired carbapenemases in Gram-negative bacteria. Clin Microbiol Infect. 2010. 16:99–101.

5. Ambretti S, Gaibani P, Caroli F, Miragliotta L, Sambri V. A carbapenem-resistant Klebsiella pneumoniae isolate harboring KPC-1 from Italy. New Microbiol. 2010. 33:281–282.
6. Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother. 2007. 51:1553–1555.
7. Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007. 51:763–765.
8. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001. 45:1151–1161.
9. Rhee JY, Park YK, Shin JY, Choi JY, Lee MY, Peck KR, et al. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob Agents Chemother. 2010. 54:2278–2279.
10. Roh KH, Lee CK, Sohn JW, Song W, Yong D, Lee K. Isolation of a Klebsiella pneumoniae isolate of sequence type 258 producing KPC-2 carbapenemase in Korea. Korean J Lab Med. 2011. 31:298–301.
11. Thomson KS. Extended-Spectrum-β-Lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010. 48:1019–1025.

12. Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009. 47:1631–1639.
13. Girlich D, Poirel L, Nordmann P. Novel ambler class A carbapenem-hydrolyzing β-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob Agents Chemother. 2010. 54:328–332.
14. Henriques I, Moura A, Alves A, Saavedra MJ, Correia A. Molecular characterization of a carbapenem-hydrolyzing class A β-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob Agents Chemother. 2004. 48:2321–2324.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download