Abstract
A slowly growing, non-chromogenic mycobacterial strain was isolated from sputum and bronchial lavage fluid samples of a patient presenting with productive cough, blood-tinged sputum, low-grade fever, and weakness. A positive acid-fast bacilli sputum smear result prompted the initiation of an anti-tuberculosis regimen. Multiplex real-time PCR showed a negative result for Mycobacterium tuberculosis complex and a positive result for nontuberculous mycobacteria. The DNA chip test confirmed this organism as a member of the genus Mycobacterium, but could not specify the species. Interestingly, the mycolic acid patterns obtained by HPLC nearly overlapped with those of M. simulans. The sequences of the Mycobacterium 16S rRNA gene and 16S-23S internal transcribed spacer region were unique and were found to have 100% similarity with those of M. riyadhense. After a review of the literature, we report this case as the first Korean case of M. riyadhense lung infection.
Currently, there are more than 125 known species of nontuberculous mycobacteria (NTM) [1, 2]. NTM are generally free-living organisms that are ubiquitous in the environment [3, 4], and are often found as contaminating organisms in laboratory or medical equipment [3, 5]. This is true especially in Korea, which is a country with a relatively high prevalence of tuberculosis (TB) [6]. NTM infection results in a disease that is not severe; however, disseminated disease may be life threatening in immunocompromised patients [3]. In recent years, NTM infections have been diagnosed in immunocompetent individuals without predisposing conditions [7, 8]. Therefore, the identification of mycobacteria that are responsible for a specific disease and the differentiation between environmental and pathogenic species are important diagnostic issues in the treatment of patients [3].
Herein, we report a case of NTM lung infection without predisposing conditions, in which an individual had been inadequately treated, thus resulting in gradual progression to chronic pulmonary disease before the consultation at our institute. In this case, the patient's condition improved only once the etiology of her disease was finally deciphered at our hospital. Of note, the recently characterized species, Mycobacterium riyadhense, was responsible for the tuberculosis-like clinical symptoms that provided our laboratory data [9, 10]. According to the literature, the following is the first case report of M. riyadhense lung infection in Korea.
A 38-yr-old woman was admitted to the pulmonology department of the Ulsan University Hospital for productive cough, blood-tinged sputum, low-grade fever, and weakness. Three months prior to her admission, she had been diagnosed with bronchiectasis at the secondary referral center but her symptoms persisted after she completed treatment there. One month before consulting our hospital, she had experienced a mild fever, weakness, and anorexia. Additionally, she was diagnosed with pneumonia. However, her condition had progressed to a more constant cough and weight loss despite previous treatment, thus she was eventually hospitalized at our institute. Upon hospitalization, a chest radiograph revealed poorly defined ground-glass opacities, which were consistent with the diagnosis of pneumonia, and a computed tomography (CT) scan showed bronchiectasis with multiple cavitary nodules.
Specimens obtained from sputum and bronchial lavage fluid revealed the presence of acid-fast bacilli, based on auramine-rhodamine-stained fluorescence microscopy. Acid-fastness was verified by Ziehl-Neelsen stained smears from colonies grown on Mycobacteria Growth Indicator Tube (MGIT; Becton Dickinson, Sparks, MD, USA) liquid medium.
Multiplex real-time PCR performed with the AdvanSureTB/NTM real-time PCR Kit (LG Lifescience, Seoul, Korea) showed a negative result for M. tuberculosis complex (MTBC) and a positive result for NTM.
Clinical and radiologic signs and symptoms of pulmonary infection including cough, fever, weight loss, and multifocal bronchiectasis with multiple small nodules and positive culture results from a single bronchial lavage fulfilled the American Thoracic Society diagnostic criteria of NTM lung disease [3]. Thus the patient was presumed to have NTM lung disease and treatment was started employing the standard regimen with isoniazid (INH), rifampicin (RIF), pyrazinamide (PZA), and ethambutol (EMB).
After a week of treatment, which was well tolerated, the general condition of the patient improved and the sputum smears became mycobacteria-negative. Therefore, the patient was discharged and advised to continue the same therapy until the final diagnosis was confirmed. Cultures grown in MGIT medium produced acid-fast bacilli in 7-9 days. In 3% Ogawa solid egg-based medium (Asan Pharmaceutical, Seoul, Korea), small, non-pigmented, smooth colonies grew in approximately 14 days at 37℃. Conventional techniques were used to test for growth and biochemical characteristics [11, 12]. The patient-derived strain UUH-10070721646 was positive for nitrate reductase, catalase, and urease, was tolerant to INH, but negative for thermotolerant catalase (Table 1). However, these phenotypic features were not sufficient to differentiate strain UUH-10070721646 from other related Mycobacteria strains.
The mycolic acid analyses were also performed using HPLC, as described previously [13]. HPLC patterns were compared with patterns from standard mycobacterial species, which were obtained from 28 ATCC standard mycobacterial species and 5 Korean Type Culture Collection (KTCC) standard mycobacterial species. The mycolic acid pattern of strain UUH-10070721646 was characterized by a single, late cluster of peaks, which was clearly distinct from TB but nearly overlapping with those of M. simulans (Fig. 1).
To identify the organism at the species level, a commercial DNA chip assay (CombiChip Mycobacteria Genotyping DNA Chip; Gene In Inc., Busan, Korea) was performed, which implements the hybridization method by using an oligonucleotide chip containing internal transcribed spacer (ITS) sequence between the 16S rRNA and 23S rRNA of Mycobacterium, thereby identifying 20 species of mycobacteria (Panmycobacteria, MTBC, M. avium-intracellular complex, M. fortuitum, M. chelonae, M. abscessus, M. kansasii, M. gordonae, M. scrofulaceum, M. szulgai, M. vaccae, M. xenopi, M. terrae, M. flavescens, M. smegmatis, M. malmoense, M. simiae, M. marinum-ulcerance, M. gastri, and M. leprae). In this case, hybridization with the genus-specific probe and the failure to hybridize with species-specific probes indicated the presence of a Mycobacterium strain that did not belong to any species that was identifiable by the system.
For complete analysis, sequencing of the 16S rRNA gene and the 16S-23S ITS region was performed with a MJ Research PTC-225 Peltier Thermal Cycler using Applied Biosystems (ABI) PRISM BigDye Terminator Cycle Sequencing Kits and ABI 3730xl sequencer (Applied Biosystems, Foster City, CA, USA) by using the standard protocol [14]. The primer pair used for amplification consisted of 27F (5'-AGA GTT TGA TC [A/C] TGG CTC AG-3') and 1492R (5'-G [C/T] T ACC TTG TTA CGA CTT-3'). This primer pair amplifies a 1,500 bp fragment of the 16S rRNA gene between positions 8 and 1509 of the Escherichia coli 16S rRNA gene. We compared the obtained sequences with the GenBank and European Molecular Biology Laboratory (EMBL; National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov) gene sequence databases. The sequencing results are list ed in Table 2. The sequences of the 16S rRNA gene and 16S-23S ITS regions of strain UUH-10070721646 were unique and closely related to the recently described species, M. riyadhense [9]. In the complete sequence of the 16S rRNA of the current isolate, the similarity to the latter species was 100%, with 1 mismatch in 1,438 bp. In the hypervariable region of hsp65 [15], there were 3 mismatches (in 423 bp; similarity 99%). In the ITS, the presence of 7 mismatches in 278 bp was responsible for a similarity of 97%. In the 744 bp stretch of rpoB, M. riyadhense presented the closest similarity (95%, with 37 mismatches). In the same regions, the similarities with M. tuberculosis were clearly lower (98% in the 16S rRNA, 86% in hsp65, 88% in the ITS, and 91% in rpoB).
The 16S rRNA gene sequence was compared with those of reference strains of the most closely related mycobacterial species present in major international nucleotide sequence databases (GenBank, EMBL, DNA Data Bank of Japan [DDBJ]) using Clustal W software version 2 (http://www.ebi.ac.uk/tools/clustalw2) [16]. The resulting topology and tree that were inferred by neighbor-joining and visualized using the Molecular Evolutionary Genetics Analysis (MEGA) software package were evaluated by bootstrap analyses based on 1,000 resamplings (Fig. 2).
Although the patient seemed to show both clinical and radiological improvement after the first regimen of INH, RIF, PZA, and EMB, INH was discontinued after 8 months of treatment due to the results of an in vitro drug susceptibility test. The drug susceptibility test was performed according to the absolute concentration method (validated for MTB strains only) in Löwenstein-Jensen medium (Green Cross Reference Laboratory, Yongin, Korea), using the first-line and second-line drugs, and the minimal inhibitory concentrations were determined using the microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) [17]. The results are interpreted following the CLSI guideline for other slowly growing NTM and newly described species, which are generally tested as for M. kansasii [17].
Strain UUH-10070721646 was found to be susceptible to RIF, EMB, kanamycin, rifabutin, amikacin, ethionamide, cycloserine, capreomycin, clarithromycin, and moxifloxacin, intermediately susceptible to ciprofloxacin, and was resistant to INH, streptomycin, ofloxacin, para-amino-salicylic acid, and levofloxacin by using the 2 methods above (Table 3).
The patient has been receiving clinical follow-up assessments for 13 months without recurrence of disease.
The incidence of pulmonary infection caused by NTM is increasing; however, it is not commonly described in Korean clinical settings. This may be explained by clinicians overlooking the possibility of an infection due to NTM, as Korea is still an endemic area for TB. Many pulmonary NTM patients are inadequately and unnecessarily treated for pulmonary TB. Furthermore, some patients are even misdiagnosed with multidrug resistant TB and treated with the secondary anti-TB regimen, as the clinical presentation of NTM is often difficult to differentiate from that of MTBC [9, 10, 18].
M. riyadhense can infect a patient without predisposing factors, resulting in the tuberculosis-like clinical symptoms that provided laboratory data from our patient. In this case, the patient's condition improved only once the etiology was finally uncovered. This is the first Korean report of a mycobacterial strain that was phenotypically and diagnostically confused with TB (but clearly distinct from it) and responsible for severe disease.
There are 4 case reports of M. riyadhense before UUH-1007 0721646 (Table 4) [9, 10, 19], the major features shared by UUH-10070721646 and these cases resulted in the confusion with MTBC. Commercial probes are frequently used for rapid identification of mycobacterial species [20]; however, M. riyadhense and other recently proposed NTM such as M. kumamotonense cross-react with MTBC DNA probes and may be overlooked by line-probe assays [18]. With the emergence of new NTM species, commercial probes could fail to discriminate between species, leaving clinical isolates either unidentified or misidentified. The clinical and radiologic signs and symptoms of pulmonary infection caused by the strain, including cough, weight loss, fever, and cavitating lung lesions, were also similar to those in typical cases caused by MTBC strains [9, 10, 19]. Another characteristic that this strain has in common with MTBC strains is the definite pathogenicity; each case showed evidence for the pathogenic role of the strain in pulmonary or extrapulmonary diseases. However, the strains differ in drug susceptibility; the first case was cured with standard anti-TB therapy of INH, RIF, and EMB that was ineffective in the second case, and the latter case was successfully treated with the combination of amikacin, ethionamide, moxifloxacin, clarithromycin, and EMB.
The strains in the third and fourth cases showed similar drug susceptibility patterns [19], which were sensitive to most first and second-line drugs, but resistant to doxicycline alone. The former was cured with INH, RIF, and EMB, while the latter patient relapsed after receiving clarithromycin and ciprofloxacin for 12 months, but then improved with anti-TB drugs (INH, RIF, EMB, PZA, clarithromycin, and ciprofloxacin). In the present case, UUH-10070721646 was treated with RIF, PZA, and EMB for 13 months without recurrence of disease.
Because of the scarcity of documented cases of M. riyadhense infection [9, 10, 19], no clinically approved agent for M. riyadhense infection is currently available [17]. Case 1 and 3 indicate that anti-TB drugs such as INH, RIF, and EMB are effective against M. riyadhense infection, but INH revealed in vitro resistance in case 2 and our case. Interestingly, the combination of clarithromycin and ciprofloxacin was not effective in case 4, despite the demonstration of in vitro susceptibility to these drugs.
Conventional laboratory culture, biochemical testing, and a limited molecular evaluation are sometimes insufficient for differentiating novel Mycobacterium species from M. tuberculosis. Biochemical methodologies are cumbersome, time-consuming, and may yield ambiguous and misleading results [21]. PCR and gene probe assays are known to yield fast and accurate results for the species-level identification of mycobacteria, but these methods are associated with the chance of contamination and a high false-positive rate, which can be difficult to sort out in various mycobacterial species and often require multiple steps to identify organisms at the species level [13, 22-24].
In the other cases of M. riyadhense, false-positive results from line-probe assays may lead to incorrect diagnoses of TB and unwarranted treatment, but in our case, misdiagnosis was prevented by PCR and HPLC, which excluded TB early on before the molecular diagnostic results were obtained. In this case, preliminary investigations by simple PCR provided a negative result, but persistent characterization of the strain by several genetic identification systems led to the first detection of M. riyadhense in Korea.
The characterization of this previously unknown pathogen raises new concerns for human health and demonstrates the continuing scope of the threat caused by NTM.
Figures and Tables
 | Fig. 1HPLC pattern of strain UUH-10070721646 (right) compared with that of Mycobacterium tuberculosis (left); the HPLC pattern of UUH-10070721646 is characterized by a single, late cluster of peaks. |
 | Fig. 2Phylogenetic relationships of strain UUH-10070721646 and related Mycobacterium species on the basis of 16S rRNA gene sequences. |
Table 1
Biochemical identification results from strain UUH-10070 721646 and related Mycobacterium species
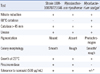
Table 3
Drug susceptibility testing pattern of strain UUH-10070721646 obtained by using the absolute concentration method in Löwenstein-Jensen medium and broth microdilution for the determination of the minimal inhibitory concentrations (MICs)

References
2. McNabb A, Eisler D, Adie K, Amos M, Rodrigues M, Stephens G, et al. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J Clin Microbiol. 2004. 42:3000–3011.
3. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of non-tuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007. 175:367–416.

4. Subcommittee of the Joint Tuberculosis Committee of the British Thoracic Society. Management of opportunist mycobacterial infections: Joint Tuberculosis Committee Guidelines 1999. Thorax. 2000. 55:210–218.
5. Forbes BA, Sahm DF, editors. Bailey and Scott's Diagnostic Microbiology. 2007. 12th ed. St. Louis: Mosby Elsevier;481.
6. Kim HJ. Current situation of tuberculosis and its control in Korea. J Korean Med Assoc. 2006. 49:762–772.

7. Prince DS, Peterson DD, Steiner RM, Gottlieb JE, Scott R, Israel HL, et al. Infection with Mycobacterium avium complex in patients without predisposing conditions. N Engl J Med. 1989. 321:863–868.
8. Arend SM, van Soolingen D, Ottenhoff TH. Diagnosis and treatment of lung infection with nontuberculous mycobacteria. Curr Opin Pulm Med. 2009. 15:201–208.

9. van Ingen J, Al-Hajoj SA, Boeree M, Al-Rabiah F, Enaimi M, de Zwaan R, et al. Mycobacterium riyadhense sp. nov., a non-tuberculous species identified as Mycobacterium tuberculosis complex by a commercial line-probe assay. Int J Syst Evol Microbiol. 2009. 59:1049–1053.
10. Tortoli E, Rogasi PG, Fantoni E, Beltrami C, De Francisci A, Mariottini A. Infection due to a novel mycobacterium, mimicking multidrug-resistant Mycobacterium tuberculosis. Clin Microbiol Infect. 2010. 16:1130–1134.
11. Lévy-Frébault VV, Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992. 42:315–323.
12. Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila ML. Separation among species of Mycobacterium terrae complex by lipid analyses: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998. 36:499–505.
13. Jeong J, Kim SR, Lee SH, Choi JI, Chang CH, Choi JY, et al. The use of High Perfomance Liquid Chromatography to speciate and characterize the epidemiology of Mycobacteria. Lab Med. 2011. 42:612–617.
14. Schuurman T, de Boer RF, Kooistra-Smid AM, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004. 42:734–740.

15. Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993. 31:175–178.

16. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2. Bioinformatics. 2007. 23:2947–2948.
17. Clinical and Laboratory Standards Institute. Susceptibility testing of Mycobacteria, Nocardia, and other aerobic actinomycetes. Approved Standard M24-A2. 2011. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute.
18. Rodríguez-Aranda A, Jimenez MS, Yubero J, Chaves F, Rubio-Garcia R, Palenque E, et al. Misidentification of Mycobacterium kumamotonense as M. tuberculosis. Emerg Infect Dis. 2010. 16:1178–1180.
19. Godreuil S, Marchandin H, Michon AL, Ponsada M, Chyderiotis G, Brisou P, et al. Mycobacterium riyadhense pulmonary infection, France and Bahrain. Emerg Infect Dis. 2012. 18:176–178.
20. Tortoli E, Nanetti A, Piersimoni C, Cichero P, Farina C, Mucignat G, et al. Performance assessment of new multiplex probe assay for identification of mycobacteria. J Clin Microbiol. 2001. 39:1079–1084.

21. Springer B, Stockman L, Teschner K, Roberts GD, Böttger EC. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996. 34:296–303.

22. Burman WJ. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis. 2000. 31:1390–1395.
23. Nah J, Huh JW, Lee SH, Kim BC, Koh YS, Pai CH. Identification of Mycobacterium tuberculosis complex using a gene probe method. Korean J Clin Pathol. 1997. 17:71–78.
24. Reisner BS, Gatson AM, Woods GL. Use of Gen-Probe AccuProbes to identify Mycobacterium avium complex, Mycobacterium tuberculosis complex, Mycobacterium kansasii, and Mycobacterium gordonae directly from BACTEC TB broth cultures. J Clin Microbiol. 1994. 32:2995–2998.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download