Abstract
Numerical and structural chromosomal abnormalities are common in hematological malignancies. Near-triploidy (58-80 chromosomes) is a numerical abnormality observed in 3% of adult cases of acute lymphoblastic leukemia. Near-triploidy is rare in myeloid lineage hematologic malignancies and compared to near-triploidy in lymphoid malignancies, near-triploidy in myeloid malignancies is associated with poor outcomes. Few studies on near-triploidy in myelodysplastic syndrome (MDS) have been reported, and the clinicopathologic significance of this condition is still unclear. Here, we report a novel case of MDS with near-triploidy and multiple structural chromosomal abnormalities: del(5q) combined with del(1p) and del(13q). These abnormalities were detected by cytogenetic analysis with array comparative genomic hybridization (CGH). Our results suggest that array CGH can be a useful tool for detecting chromosomal abnormalities in patients with MDS.
Myelodysplastic syndrome (MDS) is a heterogeneous group of clonal hematopoietic stem cell malignancies characterized by peripheral blood cytopenia(s), 1 or more lineage dysplasias, and ineffective hematopoiesis [1]. Cytogenetic abnormalities are a major predictor of prognosis in MDS. Clonal cytogenetic abnormalities have been observed in approximately 50% of de novo MDS cases [2], and in more than 80% of secondary MDS cases [3]. Near-triploidy is a cytogenetic abnormality rarely observed in MDS at the initial diagnosis. Near-triploidy has been associated with poor prognosis in MDS, but in light of the small numbers of reported cases, the clinicopathologic significance of near-triploidy in MDS is still unclear [2, 4-12].
Here, we report the case of a 68-yr-old man diagnosed with MDS/refractory cytopenia with multilineage dysplasia (RCMD) accompanied by near-triploidy with multiple structural chromosome abnormalities.
In April 2010, a 68-yr-old male patient was admitted to our hospital for the evaluation of pancytopenia and dyspnea. Physical examination revealed mild splenomegaly and swelling in both the legs. Results of peripheral blood examination were as follows: hemoglobin level, 7.7 g/dL; platelet count, 68,000/µL; and leukocyte count, 2,610/µL with 45% neutrophils, 46% lymphocytes, 7% monocytes, and 2% eosinophils. Twenty erythroblasts were observed among 100 white blood cells. The peripheral blood examination also showed the presence of anisopoikilocytic red blood cells, hypogranular platelets, and giant platelets. Bone marrow examination showed 80-90% cellular marrow with 44% (8/18) dysmegakaryopoiesis and 36% (72/200) dyserythropoiesis (including unconnected nuclei megakaryocytes, bi-nucleated cells, and nuclear budding erythroids; Fig. 1). Marrow blast cells accounted for 4% of all nucleated cells. Based on these morphological findings, the patient was diagnosed with MDS/RCMD according to the WHO classification [1]. At the time of case report preparation, the patient was discharged against medical advice, because of unidentifiable reasons. Therefore, we could not obtain more follow-up data. All methods were approved by the Institutional Review Board at Dong-A University College of Medicine, and written consent was obtained from the patient.
Cytogenetic analysis was performed on synchronized marrow cultures. The chromosomes were analyzed with the traditional G-banding method. The results of cytogenetic analysis were described according to International System for Human Cytogenetic Nomenclature (ISCN) 2009 [13]. Furthermore, we also performed array comparative genomic hybridization (CGH) analysis using DNA extracted from the marrow and the Signature OncoChip (Signature Genomics, Spokane, WA, USA). Microarray-based CGH is a high-throughput technique for studying chromosomal aberrations [14].
Of the 20 cells examined, 14 showed multiple chromosomal abnormalities; these 14 cells exhibited near-triploidy with multiple structural chromosome aberrations. The results of the bone marrow chromosome study were as follows:
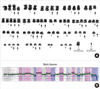
63-74,X,-X,-Y,+del(1)(p10),+3,+4,del(5)(q22q35),+6,+6,-7,+8,-9,+11,+del(13)(q12q22),+14,+14,-16,-18,-18,+19,+20,-21[cp14]/46,XY[6] (Fig. 2A).
The array CGH is shown in Fig. 2B. The results of the array CGH were as follows:
1q21.1(143,575,014-143,589,342)×1,2q34(211,992,391-213,068,103)×3,3p26.3p13(88,832-73,841,067)×3,3p13(73,877,826-74,040,678)×1,3p13q21.1(74,080,966-124,118,656)×3,3q26.1q26. 33(164,137,293-184,179,657)×3,4p16.3q35.2(45,627-191,152,793)×3,5q14.3(89,255,778-89,830,449)×1,5q21.3q33.1(105,342,555-149,489,379)×1,5q33.1q35.3(149,490,002-180,619,169)×1,6p25.3q27(128,203-170,736,131)×3,7q34(141,654,474-141,656,709)×1,7q34(141,689,169-141,720,856)×1,7q34(141,891,008-141,910,446)×1,8p23.3p23.2(177,781-3,194,314)×3, 8p23.2q24.3(3,217,735-146,263,042)×3,9p24.3q12(188,160-69,114,099)×1,9q12q34.12(69,466,291-132,741,575)×1,9q34. 12q34.3(132,743,455-140,130,559)×1,10p12.31(21,897,143-21,908,850)×3,11p15.5q25(188,204-134,425,038)×3,14q11.2q32. 33(19,528,022-105,443,403)×3,14q32.33(105,457,662-105,601,819)×3,14q32.33(105,602,180-105,638,696)×3,14q32.33(105,641,232-105,851,066)×3,14q32.33(105,896,926-106,340,244)×3,15q11.2(19,129,891-19,224,501)×3,16p13.3q24.3(35,819-88,657,641)×1,18p11.32(123,388-1,552,576)×1,18p11.31p11.22(5,509,695-9,732,828)×3,19p13.3p12(220,598-20,308,197)×3,19p12q13.43(20,633,734-63,782,017)×3,20p13q13.33(16,653-62,359,694)×3,22q11.22(21,570,725-21,579,059)×3,22q11.23(22,660,896-22,723,991)×3,Yp11.32p11.31(110,058-2,709,520)×0, Yp11.31q12(2,709,521-57,443,437)×0,Yq12(57,443,438-57,735,230)×0. Result obtained for the male patient.
The array CGH results generally corresponded with those of the chromosomal study, but a few differences are worth noting. Monosomies in chromosomes 9 and 16 were identified using CGH but were not observed in the chromosomal study. Because these monosomies were not observed while counting the chromosomes for 20 metaphase cells, these monosomies may be due to minor clonal changes. Alternatively, either chromosome of each chromosome pair may be a derivative chromosome containing chromosomal gains. Array CGH revealed cryptic aberrations, mostly copy losses on 1q, 3p, 5q, and 7q, of sizes ranging from 2.23 to 574.67 kb. However, trisomies, tetrasomies, or pentasomies were not differentiated only on the basis of array findings; Copy gains on 10q and 22q and copy losses on 18q identified by karyotypic analysis were not evident with array CGH.
Near-triploidy is rare in myeloid lineage hematologic malignancies and is associated with poor outcomes [15-17]. Further, Near-triploidy is rarely observed in MDS, and reports of near-triploidy in MDS are fewer than its reports in other myeloid malignancies. Moreover, the prognostic importance of near-triploidy in MDS is uncertain [4-12, 15]. The common cytogenetic abnormalities in MDS include -5/del(5q), -7/del(7q), and trisomy 8. Along with 5q deletion, other observed abnormalities are those affecting chromosome 17, -18/del(18q), trisomy 8, del(20q), monosomy 7, and rearrangements of chromosome 3 [18].
Lee et al. [19] reported the cytogenetic changes and prognostic features of 133 patients with MDS from Korea. About 53% of the patients with del(5q) had complex chromosome abnormalities, including aberrations reported in this study, and prognosis of these patients was mostly poor. Stamatoullas et al. [4] reported an analysis of 10 patients with MDS who showed complex hyperdiploid karyotypes. In their study, hyperdiploidy was observed in approximately 25% of the patients. Marked dyserythropoiesis was especially prevalent in cases with chromosome 13 and 17 abnormalities. Other than these findings, no significant correlations between morphologic dysplasia and chromosome abnormalities were observed. Our patient showed near-triploidy with del(5q) combined with del(1p) and del(13q), an uncommon combination of cytogenetic abnormalities in MDS.
Lim et al. [6] reported a MDS case with normal karyotype at diagnosis; however, the patient showed a secondary clonal change with massive hyperdiploidy and del(1;7)(q10;p10) as the disease progressed to acute leukemia. Our case showed near-triploidy, but there was no evidence of leukemic transformation at the time of hospitalization.
For our case, we used array CGH to confirm the cytogenetic abnormalities identified through karyotyping. Array CGH is not commonly used, but this technique offers the potential to overcome the limitations of karyotyping. Traditional karyotyping provides essential information for diagnosis and prognosis, but small aberrations may be missed during karyotyping due to the limited resolution and the requirement of dividing cells. Recently, these limitations have highlighted the utility of CGH, which is increasingly used for patients with MDS [20, 21]. In our case, array CGH detected additional copy gains in chromosomes 9 and 16; these were missed by karyotyping.
Here, we report a novel case of MDS showing near-triploidy with multiple structural chromosome abnormalities as diagnosed by cytogenetic analysis using array CGH. More studies are needed to better define the prognostic significance of near-triploidy in MDS and to evaluate the correlation between chromosomal abnormalities and patients outcomes.
Figures and Tables
Fig. 1
Bone marrow examination showed moderate dyserythropoiesis (A) and dysmegakaryopoiesis (B). Multi nucleation, nuclear budding erythroids, and unconnected nuclei megakaryocytes are marked with arrow (Wright-Giemsa stain, ×1,000).
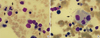
Fig. 2
(A) A representative G-banded karyotype showing massive hyperdiploidy with del(1p), del(5q), and del(13q). Chromosome numbers are listed in green, and extra chromosomes are marked with arrows. (B) Whole genomic plot of the array CGH results showing additional copy gains in chromosomes 9 and 16. The pink dots represent the patient-to-control fluorescence intensity ratio, while the blue dots show dye-reversed control-to-patient fluorescence ratios. Array CGH revealed cryptic aberrations, mostly copy losses of 2.23-574.67 kb on 1q, 3p, 5q, and 7q. However, trisomies, tetrasomies, or pentasomies were not differentiated solely on the basis of array findings, and copy gains on 10q and 22q and copy losses on 18q identified by karyotypic analysis were not observed with array CGH. See the results section for more detailed array interpretation.
Abbreviation: CGH, comparative genomic hybridization.
Acknowledgement
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MEST) (R13-2002-044-05002-0).
References
1. Sabattini E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of haematopoietic and lymphoid tissues. Pathologica. 2010. 102:83–87.
2. Solé F, Luño E, Sanzo C, Espinet B, Sanz GF, Cervera J, et al. Identification of novel cytogenetic markers with prognostic significance in a series of 968 patients with primary myelodysplastic syndromes. Haematologica. 2005. 90:1168–1178.
3. Le Beau MM, Albain KS, Larson RA, Vardiman JW, Davis EM, Blough RR, et al. Clinical and cytogenetic correlations in 63 patients with therapy-related myelodysplastic syndromes and acute nonlymphocytic leukemia: further evidence for characteristic abnormalities of chromosomes no. 5 and 7. J Clin Oncol. 1986. 4:325–345.

4. Stamatoullas A, Callat MP, Marreiros S, Tilly H, Bastard C. Unusual complex hyperdiploid karyotypes in myelodysplastic syndromes. Cancer Genet Cytogenet. 2006. 170:129–132.

5. Iyer RV, Sait SN, Matsui S, Block AW, Barcos M, Slack JL, et al. Massive hyperdiploidy and tetraploidy in acute myelocytic leukemia and myelodysplastic syndrome. Cancer Genet Cytogenet. 2004. 148:29–34.

6. Lim G, Oh SH, Choi JR, Lee HJ, Suh JT, Yoon HJ, et al. Leukemic transformation associated with massive hyperdiploidy in myelodysplastic syndrome (MDS) with der(1;7)(q10;p10): a novel case study. Leuk Res. 2010. 34:e208–e209.

7. Znoyko I, Stuart RK, Ellingham T, Winters J, Wolff DJ, Quigley DI. Tetraploidy and 5q deletion in myelodysplastic syndrome: a case report. Cancer Genet Cytogenet. 2008. 183:64–68.

8. Acar H, Calişkan U U, Kaynak M, Yildirim MS, Largaespada DA. Hyperdiploid karyotype in a childhood MDS patient. Clin Lab Haematol. 2001. 23:255–258.

9. Manley R, Cochrane J, Patton WN. Polyploidy in myelodysplastic syndrome: a case report. Cancer Genet Cytogenet. 1998. 106:170–172.
10. Tanaka M, Takeuchi H, Kaku K, Oka Y. Myelodysplastic syndrome associated with hypotriploidy. Acta Haematol. 1996. 95:148–150.

11. Bacher U, Haferlach T, Kern W, Weiss T, Schnittger S, Haferlach C. The impact of cytomorphology, cytogenetics, molecular genetics, and immunophenotyping in a comprehensive diagnostic workup of myelodysplastic syndromes. Cancer. 2009. 115:4524–4532.

12. Kawankar N, Vundinti BR. Cytogenetic abnormalities in myelodysplastic syndrome: an overview. Hematology. 2011. 16:131–138.

13. Shaffer LG, Slovak MI, Campbell LJ, editors. An International System for Human Cytogenetic Nomenclature (ISCN). 2009. Basel: S, Karger;36–84.
14. Hu J, Gao JB, Cao Y, Bottinger E, Zhang W. Exploiting noise in array CGH data to improve detection of DNA copy number change. Nucleic Acids Res. 2007. 35:e35.

15. Watanabe A, Inokuchi K, Yamaguchi H, Mizuki T, Tanosaki S, Shimada T, et al. Near-triploidy and near-tetraploidy in hematological malignancies and mutation of the p53 gene. Clin Lab Haematol. 2004. 26:25–30.

16. Sheth FJ, Sheth JJ, Desai C. Case of near triploidy with i(17)(q10) in blast crisis CML. Cancer Genet Cytogenet. 2006. 164:177–178.

17. Roland B. A case of near-triploidy in chronic myelogenous leukemia. Cancer Genet Cytogenet. 2000. 121:96–98.

18. Mallo M, Cervera J, Schanz J, Such E, García-Manero G, Luño E, et al. Impact of adjunct cytogenetic abnormalities for prognostic stratification in patients with myelodysplastic syndrome and deletion 5q. Leukemia. 2011. 25:110–120.

19. Lee HR, Oh B, Hong DS, Zang DY, Yoon HJ, Kim HJ, et al. Cytogenetic features of 5q deletion and 5q- syndrome in myelodysplastic syndrome in Korea; marker chromosomes proved to be chromosome 5 with interstitial deletion by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2010. 203:193–202.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download