Abstract
Background
Our study was to investigate the prevalence of carbapenemase genes in strains of Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in our hospital.
Methods
The carbapenemase producing Enterobacteriaceae species were confirmed by modified Hodge test (MHT) and EDTA-disc synergy test which indicating the production of class B carbapenemases. PCR and sequencing analysis were used to identify the drug-resistant genes. DNA fingerprinting based on enterobacterial repetitive intergenic consensus (ERIC)-PCR was applied to investigate the homology of Enterobacteriaceae species.
Results
From a collection of 1,472 Enterobacteriaceae species, 18 isolates with decreased susceptibility to carbapenem treatment were identified and 9 of which were positive by MHT, and 6 of which produced class B carbapenemases. PCR and sequencing analysis of the 18 isolates revealed 4 different carbapenemase genes (blaIMP-8, blaoxa-1, blaIMP-26, and blaoxa-47) in 10 isolates, with the blaIMP-8 and blaoxa-1 genes being the most common (60-70% prevalence). ERIC-PCR showed 5, 2, and 2 unique genotypes for Enterobacter cloacae, Escherichia coli, and Klebsiella pneumoniae, respectively. Three E. coli strains isolated from different patients from the urologic surgery department exhibited the same DNA banding pattern, suggesting a possible clonal dissemination. Majority (17/18) of the carbapenem-unsusceptible Enterobacteriaceae species isolates was obtained from the surgery department of our hospital.
Enterobacteriaceae species are among the most common nosocomial pathogens, causing serious infections in various organs and tissues. Currently, carbapenems are the most potent agents prescribed for the treatment of serious infections caused by Enterobacteriaceae species because of their broad spectra of antibacterial activity and their excellent stability to hydrolysis by most β-lactamases, including extended-spectrum β-lactamases (ESBLs) and AmpC cephalosporinases. However, the widespread use of carbapenems has led to the emergence of carbapenem-resistant Enterobacteriaceae species in diverse geographic locations worldwide, and this is becoming an important therapeutic challenge in the clinic setting [1-3].
The main mechanisms of carbapenem resistance in Enterobacteriaceae species include the acquisition of carbapenemases and hyperproduction of AmpC cephalosporinases, in combination with porin loss [4]. Carbapenemases are members of the molecular class A, B, and D β-lactamases, which have the ability to hydrolyze penicillins, cephalosporins, monobactams, and carbapenems [4]. Class A serine carbapenemases include 3 major families of NMC/IMI, SME, and KPC enzymes and can be inhibited by clavulanic acid and tazobactam [5]. Among the class A carbapenemases, KPC-2 is the most common type reported in China [6, 7]. Class B carbapenemases, also called metallo-β-lactamases (MBLs), are resistant to the commercially available β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam, but susceptible to inhibition by metal ion chelators, such as EDTA, a chelator of Zn2+ and other divalent cations [8]. In the past decade, a number of acquired MBLs have been identified and categorized into 2 major groups: IMP- and VIM-type enzymes. IMP-4 and IMP-8 carbapenemases have been detected in China, and these have led to a low to moderate level of carbapenem resistance in strains of Enterobacteriaceae species [9]. The hydrolysis of carbapenems by the class D oxacillinase family is weak and leads to reduced susceptibility to imipenem and meropenem but with the minimal inhibitory concentration (MIC) still in the susceptible range, thus potentially causing detection failures [10].
The goals of this study were to investigate the prevalence of carbapenemase genes in clinical strains of Enterobacteriaceae species isolated from a university hospital, and to explore the main mechanisms of decreased susceptibility to carbapenems in these clinical strains.
All patient specimens utilized in this study were from The First Affiliated Hospital of Chongqing Medical University, which has 2,500 inpatient beds and is one of the largest hospitals in the southwest of China. Samples were collected from November 2009 to December 2010. The clinical isolates were identified and the susceptibility tests were performed by using the Vitek2 Compact System with GN card and ASTGN13 card (bioMérieux, Marcy l'Etoile, France). Strains of Enterobacteriaceae species with decreased susceptibility to carbapenems (MIC of imipenem, meropenem, or ertapenem ≥2 µg/mL) were consecutively collected and confirmed by the agar dilution method, according to the guidelines of the CLSI [11].
Modified Hodge Tests (MHT) were carried out according to CLSI recommendations for phenotypic screening of carbapenemase producers among species of Enterobacteriaceae [11]. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC BAA-1705 were used as negative and positive controls, respectively. The class A and B carbapenemases were screened by clavulanic acid-disc synergy tests and EDTA-disc synergy tests, respectively, as previously described [12, 13].
Total DNA was extracted from all strains by 10 min boiling of bacterial culture, followed by 1 min centrifugation at 15,000 rpm. The supernatant was collected and used for PCR amplification. The main class A, class B, and class D carbapenemase genes were amplified using the primers and conditions described in the references listed in Table 1 [14-19]. In addition, 3 ESBL genes (blaSHV, blaTEM, and blaCTX-M) and 5 plasmid-mediated AmpC cephalosporinase genes (blaCYM-2, blaACT-1, blaACC-1, blaDHA-1, and blaFOX-1) were amplified by PCR as previously described [20, 21], and all PCR products were sequenced directly in an ABI PRISM 3730XL sequencer (Applied Biosystems, Foster City, CA, USA). DNA sequences were compared with known sequences in GenBank.
We performed genotyping on all isolates, except for Enterobacter amnigenus, by using enterobacterial repetitive intergenic consensus (ERIC)-PCR with primers ERIC1R (5'-ATGTAAGCTCCTGGGGATTCAC-3') and ERIC2 (5'-AAGTAAGTGACTGGGGTGAGCG-3') as previously described [22]. Electrophoretic banding patterns were compared by visual inspection. Isolates were considered different if their profiles differed by 2 or more bands [23].
From a collection of 1,472 isolates of Enterobacteriaceae species, we identified 18 isolates with decreased susceptibility to carbapenem treatment, including 8 isolates of Enterobacter cloacae, 5 isolates of E. coli, 4 isolates of K. pneumoniae, and 1 isolate of E. amnigenus (Table 2). The rate of non-susceptibility to carbapenems was 1.22%. Fifteen of the 18 isolates (83.3%) were resistant to ertapenem (≥8 mg/L) and only 2 of the 18 isolates (11.1%) were resistant to imipenem (≥16 mg/L). All of the isolates exhibited multidrug-resistance to the antibiotics tested (data not shown).
Of the 18 carbapenem-resistant isolates, 9 tested positive in MHT and 6 positive in the EDTA-disc synergy tests, indicating the production of class B carbapenemases in these strains (Table 2). None of them tested positive in the clavulanic acid-disc synergy tests, indicating the absence of class A carbapenemases.
Carbapenemase genes were detected in 10 of the 18 carbapenem-resistant isolates; the genes included the blaIMP-8 gene from 4 E. cloacae isolates and 2 K. pneumoniae isolates; the blaIMP-26 gene from 1 E. cloacae isolate; the blaoxa-1 gene from 5 E. cloacae isolates and 2 E. coli isolates; and the blaoxa-47 gene from 2 K. pneumoniae isolates (Table 2). We failed to detect blaSME, blaKPC, blaNDM-1, or blaVIM type carbapenemase genes in these 18 isolates. ESBL genes were found in all the isolates except for the 2 E. cloacae isolates that harbored the blaIMP-8 gene. AmpC cephalosporinase genes were detected in 7 of the 18 isolates: including the blaDHA-1 gene from 2 K. pneumoniae isolates; the blaACT-1 gene from 1 E. cloacae isolate and 1 E. amnigenus isolate; and the blaCMY-2 gene from 3 E. coli isolates. All the AmpC-positive strains, except for 1 E. cloacae strain (No. 1), did not harbor carbapenemase genes.
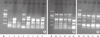
Genetic relatedness of all the isolates, except for E. amnigenus, was investigated by ERIC-PCR typing. Five distinct ERIC profiles were observed amongst 8 strains of E. cloacae, and no obvious clonal association was observed within these strains. Five strains of E. coli exhibited 2 distinct ERIC profiles, and the 3 strains presenting the same profile, i.e. showing clonal association, were all isolated from the urologic surgery department. Four strains of K. pneumoniae showed 2 ERIC profiles and an epidemiological relationship was not found (Fig. 1). However, the 2 strains with identical carbapenemase genes, ESBL and AmpC cephalosporinase, exhibited the same ERIC profile.
Since the widespread use of carbapenems in the clinic, carbapenem-resistant Enterobacteriaceae species have been detected increasingly worldwide, and a similar trend has been observed in China [9]. Effective and accurate screening for Enterobacteriaceae species with decreased susceptibility to carbapenems is important in routine clinical microbiology tests. In this study, we found that the resistance rate of ertapenem was much higher than that of imipenem (83.3% vs. 11.1%, P<0.01) in the Enterobacteriaceae species isolates studied, indicating that ertapenem was more sensitive than imipenem in the screening of carbapenem-unsusceptible Enterobacteriaceae species isolates [11]. Nine of 10 strains of Enterobacteriaceae species with carbapenemase genes tested positive in MHT, while none of the strains without any carbapenemase genes tested positive in MHT. The sensitivity and specificity of MHT were 90% and 100%, respectively. Six of 7 strains of Enterobacteriaceae species with class B carbapenemase tested positive in the EDTA-disc synergy tests, while none of the strains without class B carbapenemase genes tested positive in the EDTA-disc synergy tests. The sensitivity and specificity of the EDTA-disc synergy tests for MBLs detection were 85.7% and 100%, respectively.
In the 10 strains of Enterobacteriaceae species with carbapenemase genes, 4 strains harbored the blaIMP-8 and blaoxa-1 genes, 2 strains harbored the blaIMP-8 and blaoxa-47 genes, and 3 strains harbored the blaoxa-1 gene alone. These results suggest that the most common carbapenemase-type genes in our hospital were blaIMP-8 (6/9) and blaoxa-1 (7/9). Class A carbapenemase genes and VIM and NDM-1 β-lactamase genes were not detected in our hospital.
The blaIMP-8 gene, first found in K. pneumoniae in 2001, is very closely related to blaIMP-2 in DNA sequence, with only 4 nucleotide differences between them. This results in 2 amino acid changes. In contrast to the blaIMP-2 gene, which is located on the chromosome, the blaIMP-8 cassette is located on the plasmid, which would facilitate the spread of the resistance gene [24] and infection outbreak [25, 26]. Like blaIMP-8, class D carbapenemase can only weakly hydrolyze imipenem. All strains with blaOXA carbapenemase genes in our study were susceptible to imipenem except for one E. coli isolate (No.12), suggesting that this strain might have other drug-resistant mechanisms, such as loss or lower expression of major porins. In this study, we simultaneously detected blaIMP-8 and blaOXA-1 in 4 strains of E. cloacae. This observation has not been previously reported in China.
Biochemical analysis showed that CMY-2 and ACT-1 β-lactamase had a high catalytic efficiency towards imipenem, with low observed Km values. CMY-2, ACT-1, and DHA-1 β-lactamases conferred a high level of resistance to ceftazidime and cefotaxime and significantly reduced the susceptibility to imipenem once expressed in E. coli HB4, whilst FOX-1 and ACC-1 enzymes did not confer resistance to imipenem [21]. In our study, 6 of the 9 MHT-negative strains expressed AmpC cephalosporinase genes, including blaACT-1 in 1 E. cloacae strain, blaCMY-2 in 3 E. coli strains, and blaDHA-1 in 2 K. pneumoniae strains. The blaFOX-1 and blaACC-1 genes were not found. This observation suggests that AmpC cephalosporinase hyperproduction may contribute to the resistance observed in these isolates [4]. Five strains were not found to contain any carbapenemase or AmpC cephalosporinase genes, implying that resistance in these strains may involve other mechanisms not investigated in this study.
ERIC-PCR studies demonstrated that the infections caused by 8 strains of E. cloacae were spontaneous, because of the absence of genetic relatedness between these strains. Three of 5 E. coli strains isolated from the urologic surgery ward showed the same genotype, suggesting a possible clonal dissemination of 1 strain. Although we did not find an obvious clonal association among 4 strains of K. pneumoniae, the finding of 2 strains with completely identical ERIC profile and drug-resistant gene profile suggests there is a possibility for these to become a clonal prevalence.
Interestingly, strains expressing the blaIMP carbapenemase genes were not isolated from patients hospitalized in any department in the hospital except the department of surgery, and all the patients with positive of blaIMP carbapenemase genes had received surgery. Surgery has been reported to be an important risk factor for the acquisition of MBL producers [25]. The finding of carbapenem-resistant isolates only in the department of surgery in our hospital may have important implications for the prevention and dissemination control of these drug-resistant pathogens. Because it is likely that the strains of Enterobacteriaceae species with decreased susceptibility to carbapenems might be prevalent in the department of surgery in our hospital, strict infection control measures should be implemented in order to prevent infection dissemination.
Figures and Tables
Fig. 1
Representative gel showing banding profiles by ERIC-PCR. (A) Enterobacter cloacae; (B) Escherichia coli; (C) Klebsiella pneumoniae. The number below each lane corresponds to the strain number in Table 2. M: DNA molecular weight marker.
References
1. Oteo J, Delgado-Iribarren A, Vega D, Bautista V, Rodríguez MC, Velasco M, et al. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents. 2008. 32:534–537.

2. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother. 2006. 50:3396–3406.
3. Vatopoulos A. High rates of metallo-β-lactamase-producing Klebsiella pneumoniae in Greece--a review of the current evidence. Euro Surveill. 2008. 13:pii: 8023.
4. Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007. 20:440–458.

5. Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B, Forsman M, et al. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991. 276:269–270.

6. Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008. 52:2014–2018.
7. Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007. 51:763–765.
8. Walsh TR. The emergence and implications of metallo-β-lactamases in Gram-negative bacteria. Clin Microbiol Infect. 2005. 11:Suppl 6. 2–9.

9. Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010. 54:573–577.
10. Cuzon G, Naas T, Boqaerts P, Glupczynski Y, Huang TD, Nordmann P. Plasmid-encoded carbapenem-hydrolyzing β-lactamase OXA-48 in an imipenem-susceptible Klebsiella pneumoniae strain from Belgium. Antimicrob Agents Chemother. 2008. 52:3463–3464.
11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 21st Informational supplement, M100-S21. 2011. Wayne, PA: Clinical and Laboratory Standards Institute.
12. Radice M, Power P, Gutkind G, Fernández K, Vay C, Famiglietti A, et al. First class a carbapenemase isolated from enterobacteriaceae in Argentina. Antimicrob Agents Chemother. 2004. 48:1068–1069.
13. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum GH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001. 7:88–91.
14. Queenan AM, Torres-Viera C, Gold HS, Carmeli Y, Eliopoulos GM, Moellering RC Jr, et al. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob Agents Chemother. 2000. 44:3035–3039.
15. Bratu S, Tolaney P, Karumudi U, Quale J, Mooty M, Nichani S, et al. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J Antimicrob Chemother. 2005. 56:128–132.
16. Qi C, Malczynski M, Parker M, Scheetz MH. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J Clin Microbiol. 2008. 46:1106–1109.
17. Tsakris A, Pournaras S, Woodford N, Palepou MF, Babini GS, Douboyas J, et al. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000. 38:1290–1292.
18. Yong D, Toleman MA, Giske CG, Cho HS, Sundman S, Lee K, et al. Characterization of a new metallo-β-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009. 53:5046–5054.
19. Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004. 48:15–22.
20. Manoharan A, Premalatha K, Chatterjee S, Mathai D. SARI Group. Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011. 29:161–164.
21. Mammeri H, Guillon H, Eb F, Nordmann P. Phenotypic and biochemical. comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC β-lactamases. Antimicrob Agents Chemother. 2010. 54:4556–4560.

22. Smith JL, Drum DJ, Dai Y, Kim JM, Sanchez S, Maurer JJ, et al. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl Environ Microbiol. 2007. 73:1404–1414.
23. Khan AA, McCarthy S, Wang RF, Cerniglia CE. Characterization of United States outbreak isolates of Vibrio parahaemolyticus using enterobacterial repetitive intergenic consensus (ERIC) PCR and development of a rapid PCR method for detection of O3:K6 isolates. FEMS Microbiol Lett. 2002. 206:209–214.
24. Yan JJ, Ko WC, WU JJ. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001. 45:2368–2371.
25. Yan JJ, Ko WC, Tsai SH, Wu HM, Wu JJ. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying bla(IMP-8) in a university medical center in Taiwan. J Clin Microbiol. 2001. 39:4433–4439.
26. Yan JJ, Ko WC, Chuang CL, Wu JJ. Metallo-β-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii. J Antimicrob Chemother. 2002. 50:503–511.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download